You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002536_02345
You are here: Home > Sequence: MGYG000002536_02345
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Plesiomonas shigelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Plesiomonas; Plesiomonas shigelloides | |||||||||||
| CAZyme ID | MGYG000002536_02345 | |||||||||||
| CAZy Family | CBM73 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 158143; End: 161640 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM73 | 1113 | 1162 | 9.5e-20 | 0.9259259259259259 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13211 | PRK13211 | 9.27e-29 | 998 | 1165 | 306 | 477 | N-acetylglucosamine-binding protein GbpA. |
| smart00495 | ChtBD3 | 2.09e-07 | 1110 | 1148 | 1 | 38 | Chitin-binding domain type 3. |
| cd12214 | ChiA1_BD | 2.72e-05 | 1112 | 1161 | 1 | 45 | chitin-binding domain of Chi A1-like proteins. This group contains proteins related to the chitin binding domain of chitinase A1 (ChiA1) of Bacillus circulans WL-12. Glycosidase ChiA1 hydrolyzes chitin and is comprised of several domains: the C-terminal chitin binding domain, an N-terminal and catalytic domain, and 2 fibronectin type III-like domains. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. Bacillus circulans WL-12 ChiA1 facilitates invasion of fungal cell walls. The ChiAi chitin binding domain is required for the specific recognition of insoluble chitin. although topologically and structurally related, ChiA1 lacks the characteristic aromatic residues of Erwinia chrysanthemi endoglucanase Z (CBD(EGZ)). |
| cd12215 | ChiC_BD | 0.006 | 1111 | 1137 | 1 | 27 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIY09656.1 | 0.0 | 1 | 1165 | 2 | 1178 |
| SBT60337.1 | 0.0 | 1 | 1165 | 1 | 1177 |
| QOH80425.1 | 0.0 | 1 | 1165 | 2 | 1178 |
| QWK95135.1 | 0.0 | 1 | 1165 | 2 | 1178 |
| QWK97753.1 | 0.0 | 1 | 1165 | 2 | 1178 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A5F398 | 3.19e-17 | 832 | 982 | 848 | 995 | ToxR-activated gene A lipoprotein OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=tagA PE=3 SV=2 |
| P0C6Q7 | 1.25e-16 | 832 | 982 | 848 | 995 | ToxR-activated gene A lipoprotein OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=tagA PE=3 SV=1 |
| Q8D7V4 | 6.99e-10 | 956 | 1165 | 277 | 485 | GlcNAc-binding protein A OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=gbpA PE=3 SV=1 |
| Q7MEW9 | 9.23e-10 | 956 | 1165 | 277 | 485 | GlcNAc-binding protein A OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=gbpA PE=3 SV=1 |
| Q02I11 | 2.40e-07 | 1089 | 1165 | 309 | 389 | Chitin-binding protein CbpD OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=cpbD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
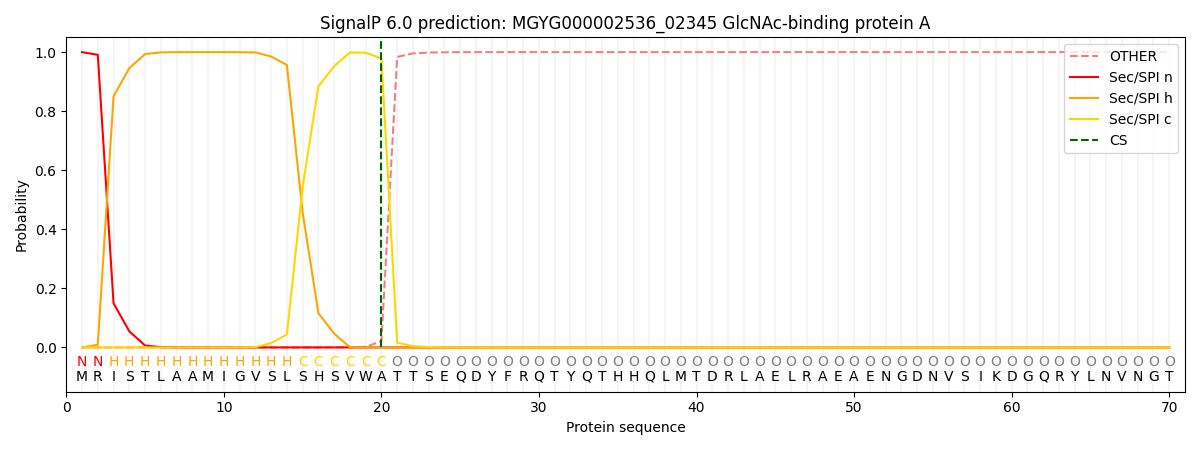
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000258 | 0.998971 | 0.000201 | 0.000191 | 0.000180 | 0.000160 |
