You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002537_00625
You are here: Home > Sequence: MGYG000002537_00625
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kocuria rhizophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Micrococcaceae; Kocuria; Kocuria rhizophila | |||||||||||
| CAZyme ID | MGYG000002537_00625 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | Extracellular serine proteinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97361; End: 98614 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04077 | Peptidases_S8_PCSK9_ProteinaseK_like | 2.63e-125 | 143 | 394 | 1 | 255 | Peptidase S8 family domain in ProteinaseK-like proteins. The peptidase S8 or Subtilase clan of proteases have a Asp/His/Ser catalytic triad that is not homologous to trypsin. This CD contains several members of this clan including: PCSK9 (Proprotein convertase subtilisin/kexin type 9), Proteinase_K, Proteinase_T, and other subtilisin-like serine proteases. PCSK9 posttranslationally regulates hepatic low-density lipoprotein receptors (LDLRs) by binding to LDLRs on the cell surface, leading to their degradation. The binding site of PCSK9 has been localized to the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR. Characterized Proteinases K are secreted endopeptidases with a high degree of sequence conservation. Proteinases K are not substrate-specific and function in a wide variety of species in different pathways. It can hydrolyze keratin and other proteins with subtilisin-like specificity. The number of calcium-binding motifs found in these differ. Proteinase T is a novel proteinase from the fungus Tritirachium album Limber. The amino acid sequence of proteinase T as deduced from the nucleotide sequence is about 56% identical to that of proteinase K. |
| cd07484 | Peptidases_S8_Thermitase_like | 1.26e-60 | 161 | 392 | 23 | 256 | Peptidase S8 family domain in Thermitase-like proteins. Thermitase is a non-specific, trypsin-related serine protease with a very high specific activity. It contains a subtilisin like domain. The tertiary structure of thermitase is similar to that of subtilisin BPN'. It contains a Asp/His/Ser catalytic triad. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) clan include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53 the it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
| cd07487 | Peptidases_S8_1 | 6.10e-60 | 165 | 393 | 1 | 264 | Peptidase S8 family domain, uncharacterized subfamily 1. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07477 | Peptidases_S8_Subtilisin_subset | 3.79e-57 | 167 | 391 | 1 | 229 | Peptidase S8 family domain in Subtilisin proteins. This group is composed of many different subtilisins: Pro-TK-subtilisin, subtilisin Carlsberg, serine protease Pb92 subtilisin, and BPN subtilisins just to name a few. Pro-TK-subtilisin is a serine protease from the hyperthermophilic archaeon Thermococcus kodakaraensis and consists of a signal peptide, a propeptide, and a mature domain. TK-subtilisin is matured from pro-TK-subtilisin upon autoprocessing and degradation of the propeptide. Unlike other subtilisins though, the folding of the unprocessed form of pro-TK-subtilisin is induced by Ca2+ binding which is almost completed prior to autoprocessing. Ca2+ is required for activity unlike the bacterial subtilisins. The propeptide is not required for folding of the mature domain unlike the bacterial subtilases because of the stability produced from Ca2+ binding. Subtilisin Carlsberg is extremely similar in structure to subtilisin BPN'/Novo thought it has a 30% difference in amino acid sequence. The substrate binding regions are also similar and 2 possible Ca2+ binding sites have been identified recently. Subtilisin Carlsberg possesses the highest commercial importance as a proteolytic additive for detergents. Serine protease Pb92, the serine protease from the alkalophilic Bacillus strain PB92, also contains two calcium ions and the overall folding of the polypeptide chain closely resembles that of the subtilisins. Members of the peptidases S8 and S35 clan include endopeptidases, exopeptidases and also a tripeptidyl-peptidase. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The S53 family contains a catalytic triad Glu/Asp/Ser. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07473 | Peptidases_S8_Subtilisin_like | 2.10e-51 | 172 | 393 | 8 | 259 | Peptidase S8 family domain in Subtilisin-like proteins. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOC24262.1 | 9.85e-108 | 50 | 408 | 24 | 377 |
| QRK11067.1 | 1.44e-94 | 101 | 409 | 90 | 404 |
| ADO75815.1 | 1.23e-90 | 47 | 408 | 42 | 397 |
| CTQ90342.1 | 6.17e-77 | 101 | 405 | 78 | 385 |
| AXO37891.1 | 6.83e-74 | 101 | 414 | 61 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WSL_A | 3.76e-102 | 136 | 408 | 1 | 274 | Structuralstudies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220],5WSL_B Structural studies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220],5WSL_C Structural studies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220] |
| 2B6N_A | 6.02e-101 | 136 | 408 | 1 | 274 | The1.8 A crystal structure of a Proteinase K like enzyme from a psychrotroph Serratia species [Serratia sp. GF96] |
| 4DZT_A | 7.96e-101 | 136 | 408 | 1 | 272 | AqualysinI: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1 [Thermus aquaticus] |
| 1S2N_A | 2.57e-93 | 138 | 415 | 1 | 276 | Crystalstructure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1S2N_B Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1SH7_A Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1SH7_B Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44] |
| 1IC6_A | 1.13e-55 | 136 | 407 | 1 | 272 | STRUCTUREOF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION [Parengyodontium album],1P7V_A Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution [Parengyodontium album],1P7W_A Crystal structure of the complex of Proteinase K with a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution [Parengyodontium album],2DQK_A Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution [Parengyodontium album],2DUJ_A Crystal structure of the complex formed between proteinase K and a synthetic peptide Leu-Leu-Phe-Asn-Asp at 1.67 A resolution [Parengyodontium album],2G4V_A anomalous substructure of proteinase K [Parengyodontium album],2HD4_A Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution [Parengyodontium album],2HPZ_A Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution [Parengyodontium album],2ID8_A Crystal structure of Proteinase K [Parengyodontium album],2PQ2_A Chain A, Proteinase K [Parengyodontium album],2PWA_A Crystal Structure of the complex of Proteinase K with Alanine Boronic acid at 0.83A resolution [Parengyodontium album],2PWB_A Crystal structure of the complex of proteinase K with coumarin at 1.9 A resolution [Parengyodontium album],2PYZ_A Crystal structure of the complex of proteinase K with auramine at 1.8A resolution [Parengyodontium album],2V8B_A SAD Structure solution of Proteinase K grown in selenate solution [Parengyodontium album],3AJ8_A X-ray analysis of Crystal of Proteinase K Obtained from H2O Solution Using PEG 8000 [Parengyodontium album],3AJ9_A X-ray analysis of Crystal of Proteinase K Obtained from D2O Solution Using PEG 8000 [Parengyodontium album],3DYB_A Chain A, Proteinase K [Parengyodontium album],3GT3_A Structure of proteinase K with the mad triangle B3C [Parengyodontium album],3GT4_A Structure of proteinase K with the magic triangle I3C [Parengyodontium album],3L1K_A SAD structure solution of proteinase K grown in potassium tellurate solution [Parengyodontium album],3OSZ_A Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution [Parengyodontium album],3Q40_A Sulphur SAD structure solution of proteinase K grown in SO4-less solution. [Parengyodontium album],3Q5G_A Sulphur SAD structure solution of proteinase K grown in SO4 solution [Parengyodontium album],3QMP_A Selenium SAD structure solution of proteinase K grown in SO4-less solution and soaked in selenate. [Parengyodontium album],4B5L_A The 1.6 A High Energy Room Temperature Structure of Proteinase K at 38.4 keV and 0.04 MGy [Parengyodontium album],4FON_A High Energy Remote SAD structure solution of Proteinase K from the 37.8 keV Tellurium K edge [Parengyodontium album],4WOB_A Proteinase-K Pre-Surface Acoustic Wave [Parengyodontium album],4WOC_A Proteinase-K Post-Surface Acoustic Waves [Parengyodontium album],4ZAR_A Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution [Parengyodontium album],5AVJ_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],5AVK_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],5B1D_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],5B1E_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],5CW1_A Proteinase K complexed with 4-iodopyrazole [Parengyodontium album],5I9S_A MicroED structure of proteinase K at 1.75 A resolution [Parengyodontium album],5KXU_A Chain A, Proteinase K [Parengyodontium album],5KXV_A Chain A, Proteinase K [Parengyodontium album],5MJL_A Chain A, Proteinase K [Parengyodontium album],5ROC_A Chain A, Proteinase K [Parengyodontium album],5ROD_A Chain A, Proteinase K [Parengyodontium album],5ROE_A Chain A, Proteinase K [Parengyodontium album],5ROF_A Chain A, Proteinase K [Parengyodontium album],5ROG_A Chain A, Proteinase K [Parengyodontium album],5ROH_A Chain A, Proteinase K [Parengyodontium album],5ROI_A Chain A, Proteinase K [Parengyodontium album],5ROJ_A Chain A, Proteinase K [Parengyodontium album],5ROK_A Chain A, Proteinase K [Parengyodontium album],5ROL_A Chain A, Proteinase K [Parengyodontium album],5ROM_A Chain A, Proteinase K [Parengyodontium album],5RON_A Chain A, Proteinase K [Parengyodontium album],5ROO_A Chain A, Proteinase K [Parengyodontium album],5ROP_A Chain A, Proteinase K [Parengyodontium album],5ROQ_A Chain A, Proteinase K [Parengyodontium album],5ROR_A Chain A, Proteinase K [Parengyodontium album],5ROS_A Chain A, Proteinase K [Parengyodontium album],5ROT_A Chain A, Proteinase K [Parengyodontium album],5ROU_A Chain A, Proteinase K [Parengyodontium album],5ROV_A Chain A, Proteinase K [Parengyodontium album],5ROW_A Chain A, Proteinase K [Parengyodontium album],5ROX_A Chain A, Proteinase K [Parengyodontium album],5ROY_A Chain A, Proteinase K [Parengyodontium album],5ROZ_A Chain A, Proteinase K [Parengyodontium album],5RP0_A Chain A, Proteinase K [Parengyodontium album],5RP1_A Chain A, Proteinase K [Parengyodontium album],5RP2_A Chain A, Proteinase K [Parengyodontium album],5RP3_A Chain A, Proteinase K [Parengyodontium album],5RP4_A Chain A, Proteinase K [Parengyodontium album],5RP5_A Chain A, Proteinase K [Parengyodontium album],5RP6_A Chain A, Proteinase K [Parengyodontium album],5RP7_A Chain A, Proteinase K [Parengyodontium album],5RP8_A Chain A, Proteinase K [Parengyodontium album],5RP9_A Chain A, Proteinase K [Parengyodontium album],5RPA_A Chain A, Proteinase K [Parengyodontium album],5RPB_A Chain A, Proteinase K [Parengyodontium album],5RPC_A Chain A, Proteinase K [Parengyodontium album],5RPD_A Chain A, Proteinase K [Parengyodontium album],5RPE_A Chain A, Proteinase K [Parengyodontium album],5RPF_A Chain A, Proteinase K [Parengyodontium album],5RPG_A Chain A, Proteinase K [Parengyodontium album],5RPH_A Chain A, Proteinase K [Parengyodontium album],5RPI_A Chain A, Proteinase K [Parengyodontium album],5RPJ_A Chain A, Proteinase K [Parengyodontium album],5RPK_A Chain A, Proteinase K [Parengyodontium album],5RPL_A Chain A, Proteinase K [Parengyodontium album],5RPM_A Chain A, Proteinase K [Parengyodontium album],5RPN_A Chain A, Proteinase K [Parengyodontium album],5RPO_A Chain A, Proteinase K [Parengyodontium album],5RPP_A Chain A, Proteinase K [Parengyodontium album],5RPQ_A Chain A, Proteinase K [Parengyodontium album],5RPR_A Chain A, Proteinase K [Parengyodontium album],5RPS_A Chain A, Proteinase K [Parengyodontium album],5RPT_A Chain A, Proteinase K [Parengyodontium album],5RPU_A Chain A, Proteinase K [Parengyodontium album],5RPV_A Chain A, Proteinase K [Parengyodontium album],5RPW_A Chain A, Proteinase K [Parengyodontium album],5RPX_A Chain A, Proteinase K [Parengyodontium album],5RPY_A Chain A, Proteinase K [Parengyodontium album],5RPZ_A Chain A, Proteinase K [Parengyodontium album],5UVL_A Chain A, Proteinase K [Parengyodontium album],5WHW_A Using sound pulses to solve the crystal harvesting bottleneck [Parengyodontium album],5WJG_A Using sound pulses to solve the crystal harvesting bottleneck [Parengyodontium album],5WJH_A Using sound pulses to solve the crystal harvesting bottleneck [Parengyodontium album],5WRC_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6FJS_A Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data [Parengyodontium album],6J43_A Chain A, Proteinase K [Parengyodontium album],6K2P_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2R_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2S_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2T_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2V_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2W_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6K2X_A Crystal structure of proteinase K from Engyodontium album [Parengyodontium album],6LAW_A MicroED structure of proteinase K at 1.50A determained using crystal lamellas prepared by focused ion beam milling [Parengyodontium album],6MH6_A High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. [Parengyodontium album],6N4U_A MicroED structure of Proteinase K at 2.75A resolution from a single milled crystal. [Parengyodontium album],6QF1_A Chain A, Proteinase K [Parengyodontium album],6QXV_A Pink beam serial crystallography: Proteinase K, 1 us exposure, 1585 patterns merged (2 chips) [Parengyodontium album],6RUG_A Co-substituted alpha-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RUH_A Ni-substituted alpha-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RUK_A Cu-substituted alpha-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RUN_A Co-substituted alpha-Keggin bound to Proteinase K solved by EP [Parengyodontium album],6RUW_A Zn-substituted alpha-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RVE_A Co-substituted beta-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RVG_A Co-substituted beta-Keggin bound to Proteinase K solved by MR [Parengyodontium album],6RZP_A Multicrystal structure of Proteinase K at room temperature using a multilayer monochromator. [Parengyodontium album],6TXG_A Proteinase K in complex with a 'half sandwich'-type Ru(II) coordination compound [Parengyodontium album],6V8R_A Proteinase K Determined by MicroED Phased by ARCIMBOLDO_SHREDDER [Parengyodontium album],6ZET_AAA Chain AAA, Proteinase K [Parengyodontium album],6ZEU_AAA Chain AAA, Proteinase K [Parengyodontium album],6ZEV_AAA Chain AAA, Proteinase K [Parengyodontium album],7A68_A proteinase K crystallized from 0.5 M NaNO3 [Parengyodontium album],7A9F_A Chain A, Proteinase K [Parengyodontium album],7A9K_A Chain A, Proteinase K [Parengyodontium album],7A9M_A Chain A, Proteinase K [Parengyodontium album],7C0P_A Chain A, Proteinase K [Parengyodontium album],7LN7_A Chain A, Proteinase K [Parengyodontium album],7LPT_A Chain A, Proteinase K [Parengyodontium album],7LPU_A Chain A, Proteinase K [Parengyodontium album],7LPV_A Chain A, Proteinase K [Parengyodontium album],7LQ8_A Chain A, Proteinase K [Parengyodontium album],7LQ9_A Chain A, Proteinase K [Parengyodontium album],7LQA_A Chain A, Proteinase K [Parengyodontium album],7LQB_A Chain A, Proteinase K [Parengyodontium album],7LQC_A Chain A, Proteinase K [Parengyodontium album],7NJJ_A Chain A, Proteinase K [Parengyodontium album],7NUY_A Chain A, Proteinase K [Parengyodontium album],7NUZ_A Chain A, Proteinase K [Parengyodontium album],7S4Z_A Chain A, Proteinase K [Parengyodontium album] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P80146 | 2.18e-107 | 28 | 410 | 22 | 408 | Extracellular serine proteinase OS=Thermus sp. (strain Rt41A) OX=32063 PE=1 SV=3 |
| P08594 | 9.91e-106 | 92 | 416 | 83 | 407 | Aqualysin-1 OS=Thermus aquaticus OX=271 GN=pstI PE=1 SV=2 |
| P16588 | 4.53e-103 | 63 | 408 | 63 | 412 | Alkaline serine exoprotease A OS=Vibrio alginolyticus OX=663 GN=proA PE=3 SV=1 |
| G1X8P8 | 6.13e-73 | 50 | 405 | 39 | 401 | Cuticle-degrading serine protease OS=Arthrobotrys oligospora (strain ATCC 24927 / CBS 115.81 / DSM 1491) OX=756982 PE=1 SV=1 |
| P40903 | 2.04e-72 | 26 | 388 | 64 | 438 | Sexual differentiation process putative subtilase-type proteinase isp6 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=isp6 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
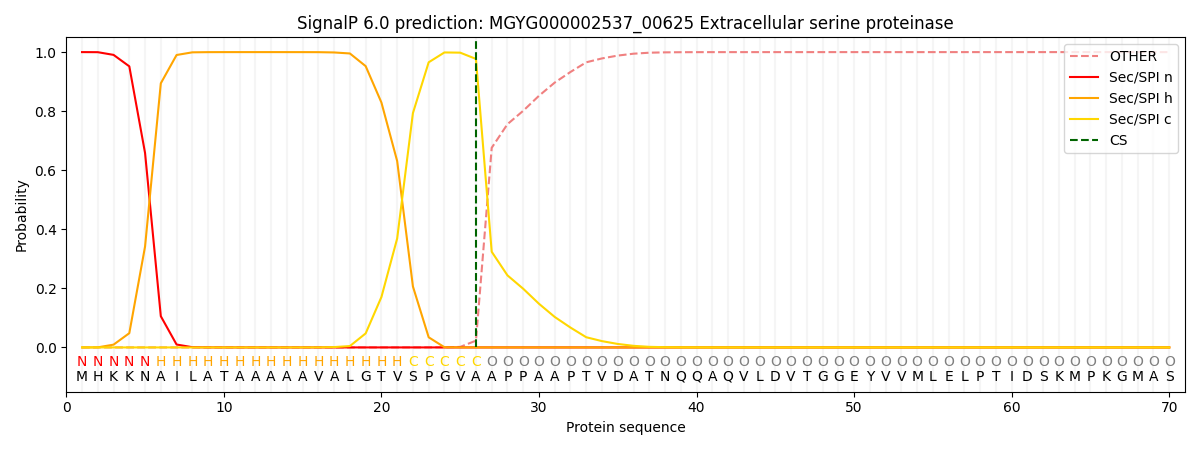
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.998890 | 0.000186 | 0.000230 | 0.000197 | 0.000160 |
