You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002548_02164
You are here: Home > Sequence: MGYG000002548_02164
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
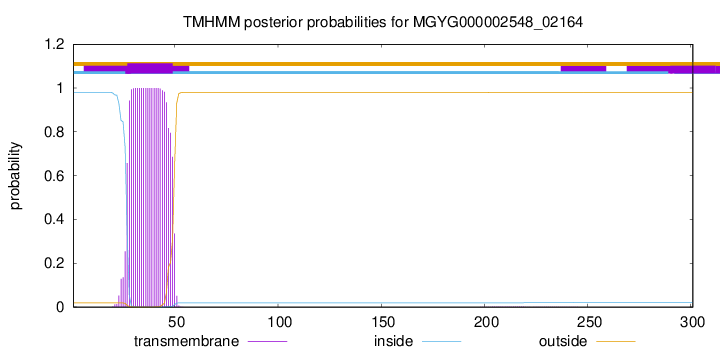
TMHMM annotations
Basic Information help
| Species | Staphylococcus argenteus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus argenteus | |||||||||||
| CAZyme ID | MGYG000002548_02164 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21923; End: 22828 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 86 | 272 | 3.6e-52 | 0.9661016949152542 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 5.02e-74 | 77 | 292 | 58 | 260 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 2.03e-69 | 82 | 273 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG5009 | MrcA | 5.17e-49 | 49 | 288 | 26 | 246 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 8.24e-36 | 105 | 286 | 76 | 243 | penicillin-binding protein 1a; Provisional |
| COG4953 | PbpC | 2.29e-35 | 104 | 287 | 67 | 235 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATY57280.1 | 1.28e-201 | 1 | 301 | 1 | 301 |
| BBD86513.1 | 1.28e-201 | 1 | 301 | 1 | 301 |
| BBN29416.1 | 1.28e-201 | 1 | 301 | 1 | 301 |
| ATZ87502.1 | 1.28e-201 | 1 | 301 | 1 | 301 |
| CCE59421.1 | 7.41e-201 | 1 | 301 | 1 | 301 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JE5_A | 1.86e-47 | 63 | 296 | 22 | 255 | StructuralAnd Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6],2JE5_B Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6] |
| 5U2G_A | 1.33e-26 | 107 | 291 | 52 | 222 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 3NB6_A | 1.77e-26 | 106 | 287 | 31 | 198 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 3UDF_A | 4.20e-26 | 85 | 298 | 30 | 228 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 7U4H_A | 4.59e-26 | 72 | 291 | 18 | 247 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31776 | 1.19e-26 | 107 | 291 | 67 | 237 | Penicillin-binding protein 1A OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=mrcA PE=1 SV=2 |
| A7FY32 | 2.15e-26 | 92 | 288 | 73 | 264 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 2.15e-26 | 92 | 288 | 73 | 264 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 2.15e-26 | 92 | 288 | 73 | 264 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| P02918 | 5.39e-26 | 105 | 286 | 76 | 243 | Penicillin-binding protein 1A OS=Escherichia coli (strain K12) OX=83333 GN=mrcA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
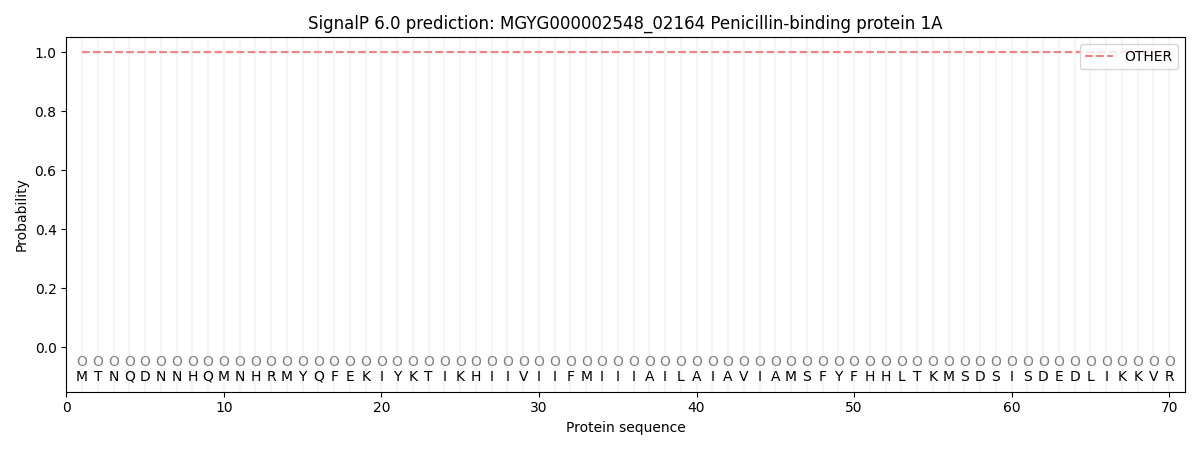
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999980 | 0.000049 | 0.000000 | 0.000000 | 0.000000 | 0.000002 |