You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002575_00456
You are here: Home > Sequence: MGYG000002575_00456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
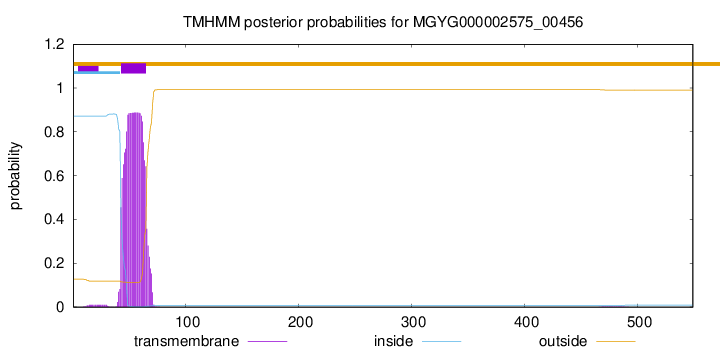
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; ; | |||||||||||
| CAZyme ID | MGYG000002575_00456 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10; End: 1659 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 4.16e-16 | 8 | 99 | 221 | 312 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 5.16e-06 | 12 | 89 | 250 | 325 | beta-galactosidase; Provisional |
| PLN03059 | PLN03059 | 9.73e-05 | 453 | 498 | 620 | 661 | beta-galactosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUI47871.1 | 2.00e-295 | 1 | 549 | 279 | 829 |
| ANQ59332.1 | 1.63e-294 | 1 | 549 | 279 | 829 |
| CUA17207.1 | 1.63e-294 | 1 | 549 | 279 | 829 |
| QUU05629.1 | 6.57e-294 | 1 | 549 | 279 | 829 |
| QIH32971.1 | 1.75e-274 | 1 | 549 | 289 | 838 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EON_A | 8.65e-09 | 401 | 532 | 479 | 617 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
| 3D3A_A | 5.35e-08 | 483 | 532 | 533 | 597 | Crystalstructure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49676 | 1.61e-08 | 4 | 498 | 239 | 665 | Beta-galactosidase OS=Brassica oleracea OX=3712 PE=2 SV=1 |
| Q9FN08 | 2.02e-08 | 4 | 509 | 244 | 677 | Beta-galactosidase 10 OS=Arabidopsis thaliana OX=3702 GN=BGAL10 PE=2 SV=1 |
| Q0INM3 | 3.82e-08 | 4 | 510 | 276 | 742 | Beta-galactosidase 15 OS=Oryza sativa subsp. japonica OX=39947 GN=Os12g0429200 PE=2 SV=1 |
| A1D199 | 9.00e-08 | 4 | 221 | 295 | 522 | Probable beta-galactosidase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=lacB PE=3 SV=1 |
| Q5BEQ0 | 2.06e-07 | 4 | 106 | 295 | 398 | Probable beta-galactosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=lacB PE=3 SV=2 |
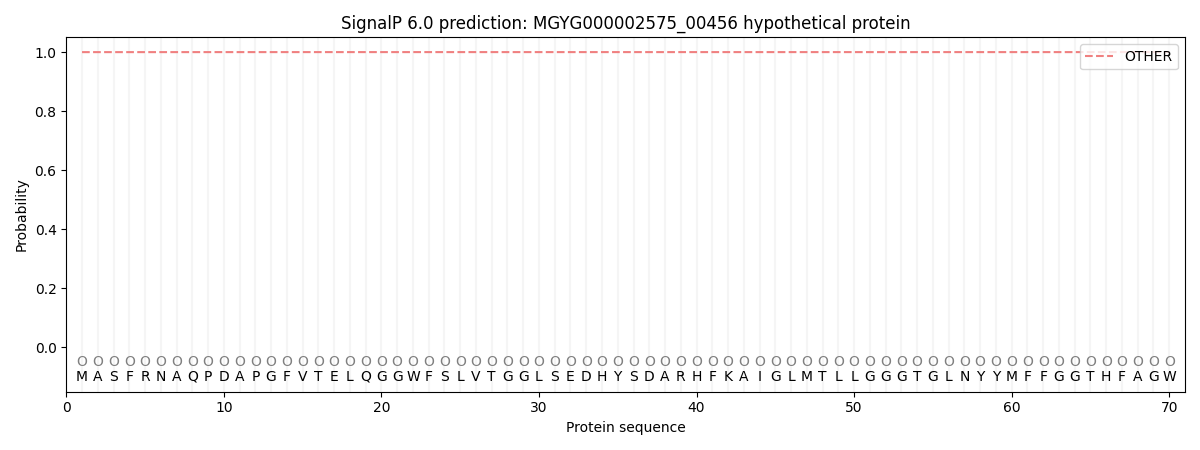
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999966 | 0.000034 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |