You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002584_00746
You are here: Home > Sequence: MGYG000002584_00746
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900545155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900545155 | |||||||||||
| CAZyme ID | MGYG000002584_00746 | |||||||||||
| CAZy Family | GH63 | |||||||||||
| CAZyme Description | Cytoplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 180372; End: 182072 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01204 | Trehalase | 3.67e-21 | 305 | 479 | 326 | 497 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| PRK10137 | PRK10137 | 8.20e-18 | 305 | 434 | 599 | 744 | alpha-glucosidase; Provisional |
| pfam03200 | Glyco_hydro_63 | 9.68e-16 | 146 | 493 | 178 | 493 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| COG1626 | TreA | 5.38e-15 | 70 | 435 | 157 | 498 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PRK13270 | treF | 8.00e-11 | 297 | 463 | 362 | 518 | alpha,alpha-trehalase TreF. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAT91813.1 | 2.94e-188 | 33 | 565 | 35 | 507 |
| QWP09187.1 | 4.02e-188 | 33 | 565 | 4 | 476 |
| QWP15489.1 | 4.02e-188 | 33 | 565 | 4 | 476 |
| QWP35628.1 | 4.02e-188 | 33 | 565 | 4 | 476 |
| QWP56518.1 | 4.02e-188 | 33 | 565 | 4 | 476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W7S_A | 4.72e-08 | 251 | 438 | 522 | 714 | Escherichiacoli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7S_B Escherichia coli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7T_A Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7T_B Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7U_A Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12],3W7U_B Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12] |
| 3W7W_A | 4.72e-08 | 251 | 438 | 522 | 714 | Crystalstructure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],3W7W_B Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],5GW7_A Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5GW7_B Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 3W7X_A | 4.72e-08 | 251 | 438 | 522 | 714 | Crystalstructure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],3W7X_B Crystal structure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],5CA3_A Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5CA3_B Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 6XUX_A | 5.03e-08 | 251 | 438 | 71 | 263 | ChainA, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody [Escherichia coli K-12] |
| 7PQQ_B | 5.03e-08 | 251 | 438 | 71 | 263 | ChainB, Anti-RON nanobody,Megabody 91,Glucosidase YgjK [Lama glama] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94250 | 9.41e-20 | 284 | 508 | 180 | 407 | Uncharacterized protein BB_0381 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0381 PE=4 SV=2 |
| O42783 | 5.87e-10 | 303 | 433 | 513 | 645 | Cytosolic neutral trehalase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=treB PE=2 SV=2 |
| J5K1E2 | 8.50e-08 | 305 | 433 | 514 | 644 | Cytosolic neutral trehalase OS=Beauveria bassiana (strain ARSEF 2860) OX=655819 GN=NTH1 PE=1 SV=1 |
| C0Q162 | 9.49e-08 | 297 | 479 | 362 | 535 | Cytoplasmic trehalase OS=Salmonella paratyphi C (strain RKS4594) OX=476213 GN=treF PE=3 SV=1 |
| Q8ZLC8 | 9.49e-08 | 297 | 479 | 362 | 535 | Cytoplasmic trehalase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=treF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
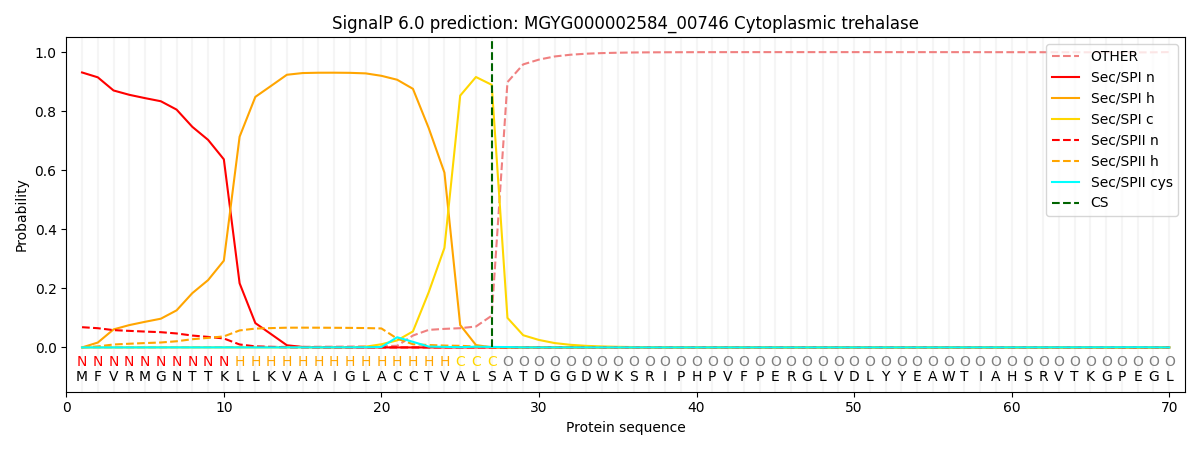
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001137 | 0.925105 | 0.072949 | 0.000304 | 0.000253 | 0.000229 |
