You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002603_01838
You are here: Home > Sequence: MGYG000002603_01838
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900767615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900767615 | |||||||||||
| CAZyme ID | MGYG000002603_01838 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41557; End: 43746 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 449 | 713 | 7.8e-46 | 0.6831683168316832 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 4.01e-31 | 438 | 715 | 89 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.87e-28 | 452 | 710 | 54 | 260 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 9.13e-18 | 459 | 693 | 126 | 321 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 2.50e-11 | 46 | 133 | 1 | 87 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.71e-07 | 74 | 133 | 51 | 110 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67590.1 | 6.99e-271 | 13 | 726 | 16 | 732 |
| QRQ48288.1 | 8.95e-240 | 1 | 717 | 2 | 716 |
| QUT46089.1 | 8.27e-238 | 1 | 717 | 2 | 716 |
| QIU93949.1 | 4.24e-212 | 4 | 717 | 6 | 725 |
| QDM11106.1 | 1.32e-210 | 4 | 717 | 6 | 725 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 8.83e-18 | 444 | 710 | 114 | 335 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 4MGS_A | 1.99e-16 | 148 | 296 | 1 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4W8L_A | 8.88e-14 | 453 | 724 | 106 | 352 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 1W32_A | 4.89e-13 | 464 | 684 | 109 | 310 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W3H_A | 5.35e-13 | 464 | 684 | 120 | 321 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W3H_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q96VB6 | 4.46e-15 | 453 | 710 | 123 | 316 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| O94163 | 4.92e-14 | 453 | 710 | 125 | 320 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| W0HFK8 | 1.66e-13 | 453 | 717 | 131 | 331 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| G4MTF8 | 3.99e-13 | 453 | 710 | 129 | 323 | Endo-1,4-beta-xylanase 2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL2 PE=3 SV=1 |
| O69230 | 4.97e-13 | 453 | 725 | 472 | 719 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
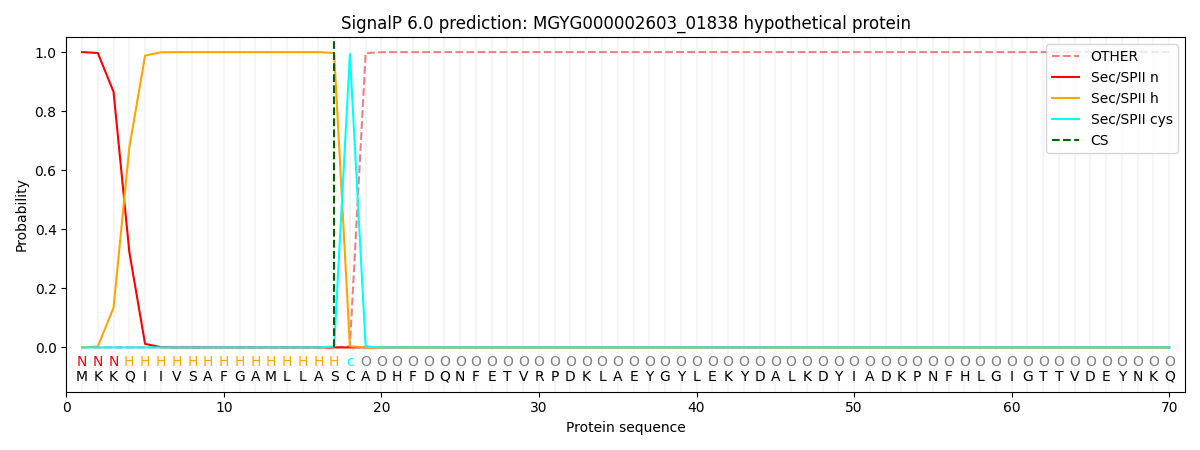
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000018 | 0.000000 | 0.000000 | 0.000000 |
