You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002621_00292
You are here: Home > Sequence: MGYG000002621_00292
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides ndongoniae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ndongoniae | |||||||||||
| CAZyme ID | MGYG000002621_00292 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 151502; End: 152395 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 150 | 280 | 4.1e-17 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 8.75e-16 | 148 | 279 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.83e-07 | 144 | 232 | 9 | 92 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP56724.1 | 6.12e-126 | 1 | 297 | 1 | 297 |
| QQT78032.1 | 6.12e-126 | 1 | 297 | 1 | 297 |
| QUU07363.1 | 6.12e-126 | 1 | 297 | 1 | 297 |
| QUT89968.1 | 4.12e-122 | 1 | 297 | 1 | 299 |
| ALK86788.1 | 7.54e-95 | 1 | 297 | 1 | 296 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2J7M_A | 3.70e-07 | 141 | 227 | 12 | 92 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 3.77e-07 | 141 | 227 | 13 | 93 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 6.04e-07 | 93 | 227 | 546 | 680 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 3.55e-06 | 93 | 227 | 576 | 710 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 3.55e-06 | 93 | 227 | 576 | 710 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
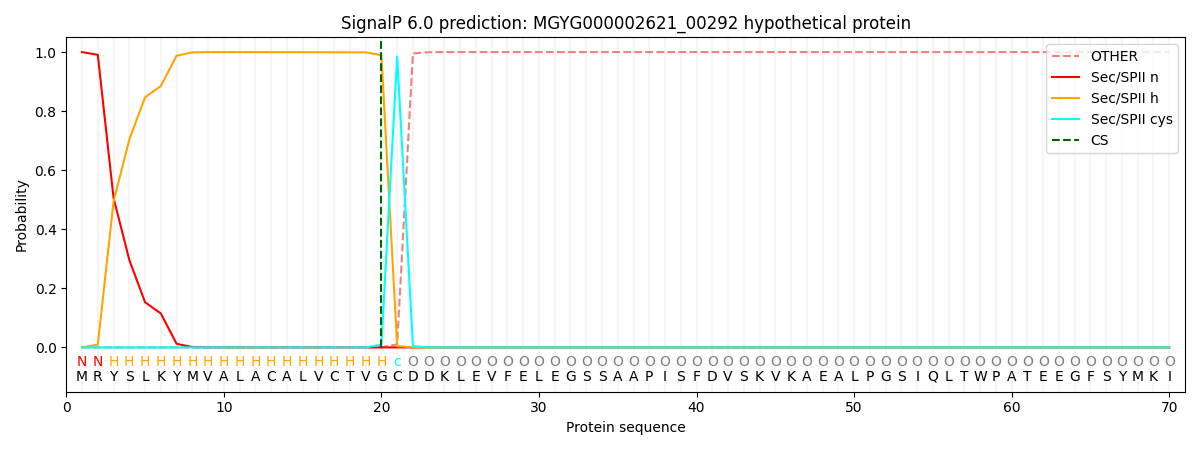
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000019 | 0.000000 | 0.000000 | 0.000000 |
