You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002621_01936
You are here: Home > Sequence: MGYG000002621_01936
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides ndongoniae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ndongoniae | |||||||||||
| CAZyme ID | MGYG000002621_01936 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51826; End: 53595 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 264 | 554 | 2.4e-83 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 8.34e-43 | 258 | 552 | 10 | 267 | Cellulase (glycosyl hydrolase family 5). |
| pfam16389 | DUF4998 | 2.06e-26 | 21 | 219 | 1 | 199 | Domain of unknown function. This family around 200 residues locates in the N-terminal of some uncharacterized proteins in various Bacteroides and Parabacteroides species. The function of this family remains unknown. |
| COG2730 | BglC | 4.79e-20 | 237 | 557 | 40 | 362 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY45078.1 | 1.71e-98 | 183 | 589 | 75 | 492 |
| QRM99680.1 | 2.80e-89 | 214 | 554 | 185 | 542 |
| QEL01247.1 | 2.43e-86 | 230 | 589 | 45 | 409 |
| QBJ18684.1 | 1.66e-84 | 235 | 589 | 221 | 575 |
| QPH58994.1 | 1.66e-84 | 235 | 589 | 221 | 575 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4W8A_A | 6.98e-68 | 239 | 589 | 1 | 378 | Crystalstructure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form [uncultured bacterium],4W8B_A Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in complex with XXLG [uncultured bacterium] |
| 5OYC_A | 1.29e-67 | 232 | 584 | 36 | 395 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6HA9_A | 9.67e-67 | 232 | 584 | 36 | 395 | Structureof an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HA9_B Structure of an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_A Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_B Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107] |
| 2JEP_A | 2.67e-52 | 240 | 583 | 34 | 395 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6PZ7_A | 2.23e-48 | 243 | 564 | 11 | 321 | GH5-4broad specificity endoglucanase from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 3.18e-51 | 241 | 583 | 40 | 400 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P28623 | 2.67e-44 | 241 | 556 | 43 | 341 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P17901 | 1.24e-43 | 213 | 566 | 24 | 386 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P23660 | 2.28e-43 | 240 | 565 | 26 | 348 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P10477 | 2.30e-43 | 240 | 558 | 56 | 356 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
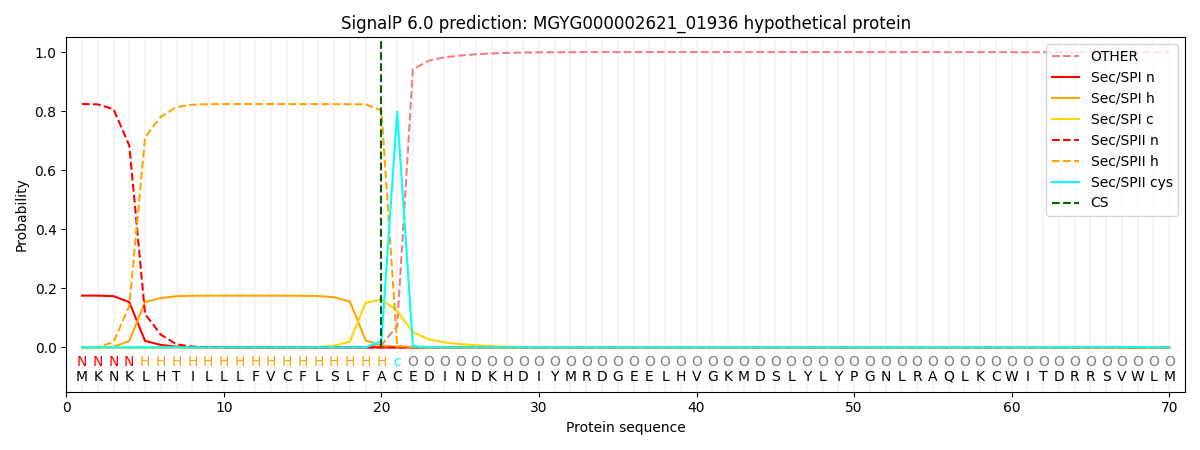
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000320 | 0.171163 | 0.828252 | 0.000091 | 0.000097 | 0.000083 |
