You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002622_01137
You are here: Home > Sequence: MGYG000002622_01137
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900546095 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900546095 | |||||||||||
| CAZyme ID | MGYG000002622_01137 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 44987; End: 46615 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 291 | 519 | 8.4e-30 | 0.793918918918919 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00704 | Glyco_hydro_18 | 7.22e-25 | 310 | 516 | 65 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 2.65e-21 | 299 | 516 | 57 | 334 | Glyco_18 domain. |
| cd06548 | GH18_chitinase | 5.92e-21 | 319 | 516 | 97 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd06545 | GH18_3CO4_chitinase | 1.22e-15 | 328 | 529 | 80 | 253 | The Bacteroides thetaiotaomicron protein represented by pdb structure 3CO4 is an uncharacterized bacterial member of the family 18 glycosyl hydrolases with homologs found in Flavobacterium, Stigmatella, and Pseudomonas. |
| cd02879 | GH18_plant_chitinase_class_V | 1.09e-13 | 288 | 516 | 46 | 293 | The class V plant chitinases have a glycosyl hydrolase family 18 (GH18) domain, but lack the chitin-binding domain present in other GH18 enzymes. The GH18 domain of the class V chitinases has endochitinase activity in some cases and no catalytic activity in others. Included in this family is a lectin found in black locust (Robinia pseudoacacia) bark, which binds chitin but lacks chitinase activity. Also included is a chitinase-related receptor-like kinase (CHRK1) from tobacco (Nicotiana tabacum), with an N-terminal GH18 domain and a C-terminal kinase domain, which is thought to be part of a plant signaling pathway. The GH18 domain of CHRK1 is expressed extracellularly where it binds chitin but lacks chitinase activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBJ17915.1 | 7.29e-132 | 20 | 534 | 300 | 815 |
| QGT71432.1 | 1.85e-128 | 22 | 534 | 37 | 539 |
| QDM09081.1 | 1.85e-128 | 22 | 534 | 37 | 539 |
| QUT80683.1 | 1.85e-128 | 22 | 534 | 37 | 539 |
| QUT24911.1 | 8.29e-124 | 22 | 534 | 37 | 539 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XYZ_A | 9.47e-16 | 322 | 532 | 94 | 304 | Crystalstructure of the GH18 chitinase ChiB from the chitin utilization locus of Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101] |
| 3AQU_A | 7.82e-10 | 308 | 531 | 66 | 341 | Crystalstructure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_B Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_C Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_D Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana] |
| 5XWQ_A | 1.53e-09 | 271 | 536 | 11 | 372 | Crystalstructure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) [Rhizomucor miehei],5YUQ_A Chain A, Chintase [Rhizomucor miehei],5YUQ_B Chain B, Chintase [Rhizomucor miehei],7FBT_A Chain A, Chitinase [Rhizomucor miehei] |
| 5XWF_A | 4.77e-09 | 271 | 518 | 11 | 313 | ChainA, Fungal chitinase from Rhizomucor miehei (SeMet-substituted proteins) [Rhizomucor miehei] |
| 1ITX_A | 2.23e-08 | 299 | 525 | 113 | 415 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O81862 | 4.76e-09 | 308 | 531 | 89 | 364 | Class V chitinase OS=Arabidopsis thaliana OX=3702 GN=ChiC PE=1 SV=1 |
| P20533 | 1.76e-07 | 299 | 525 | 145 | 447 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| A0A1B1J8Z2 | 2.02e-07 | 290 | 520 | 97 | 386 | Class V chitinase CHIT5 OS=Lotus japonicus OX=34305 GN=CHIT5 PE=1 SV=2 |
| A6N6J0 | 8.98e-07 | 247 | 521 | 55 | 395 | Endochitinase 46 OS=Trichoderma harzianum OX=5544 GN=chit46 PE=1 SV=1 |
| P48827 | 2.05e-06 | 247 | 521 | 54 | 388 | Endochitinase 42 OS=Trichoderma harzianum OX=5544 GN=chit42 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
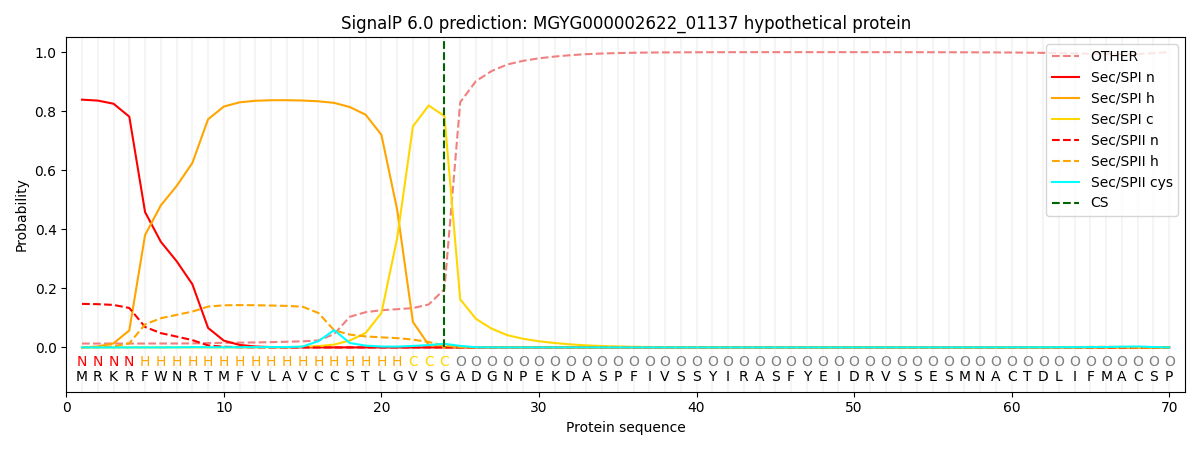
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.019985 | 0.822967 | 0.154448 | 0.001464 | 0.000562 | 0.000533 |
