You are browsing environment: HUMAN GUT
MGYG000002722_00050
Basic Information
help
Species
UBA1820 sp002314265
Lineage
Bacteria; Bacteroidota; Bacteroidia; Flavobacteriales; UBA1820; UBA1820; UBA1820 sp002314265
CAZyme ID
MGYG000002722_00050
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002722
1851778
MAG
Canada
North America
Gene Location
Start: 51406;
End: 53019
Strand: +
No EC number prediction in MGYG000002722_00050.
Family
Start
End
Evalue
family coverage
GT83
8
438
2.4e-63
0.8111111111111111
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
1.12e-23
16
421
23
426
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
pfam13231
PMT_2
2.29e-06
56
212
1
151
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366.
more
PRK13279
arnT
2.68e-06
25
417
31
429
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
1.02e-09
17
438
48
476
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
2.62e-13
10
334
5
328
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
A8GDR9
7.14e-09
23
361
30
371
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Serratia proteamaculans (strain 568) OX=399741 GN=arnT PE=3 SV=1
more
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.104828
0.891955
0.001125
0.000676
0.000628
0.000745
start
end
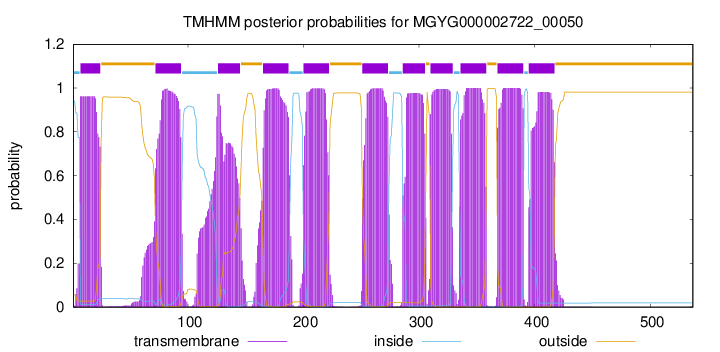
7
24
72
94
126
145
165
187
200
222
251
273
286
305
310
329
336
358
368
390
395
417