You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002737_00831
You are here: Home > Sequence: MGYG000002737_00831
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900553465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900553465 | |||||||||||
| CAZyme ID | MGYG000002737_00831 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15585; End: 16991 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 36 | 445 | 2.7e-123 | 0.9955156950672646 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01532 | Glyco_hydro_47 | 7.27e-140 | 36 | 445 | 1 | 452 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| PTZ00470 | PTZ00470 | 4.72e-115 | 19 | 445 | 65 | 517 | glycoside hydrolase family 47 protein; Provisional |
| COG1331 | YyaL | 5.50e-05 | 116 | 210 | 472 | 570 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANQ52185.2 | 1.98e-235 | 22 | 455 | 41 | 479 |
| QWG04411.1 | 1.98e-235 | 22 | 455 | 41 | 479 |
| AZQ63854.1 | 3.51e-229 | 22 | 455 | 42 | 480 |
| QWG10463.1 | 6.35e-229 | 22 | 455 | 39 | 477 |
| AIM36873.1 | 8.58e-212 | 12 | 448 | 5 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4AYO_A | 1.12e-70 | 27 | 445 | 11 | 434 | Structureof The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 [Caulobacter sp. K31],4AYP_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside [Caulobacter sp. K31],4AYQ_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with mannoimidazole [Caulobacter sp. K31],4AYR_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin [Caulobacter sp. K31],5MEH_A Crystal structure of alpha-1,2-mannosidase from Caulobacter K31 strain in complex with 1-deoxymannojirimycin [Caulobacter sp. K31],5NE5_A Crystal structure of family 47 alpha-1,2-mannosidase from Caulobacter K31 strain in complex with kifunensine [Caulobacter sp. K31] |
| 5KIJ_A | 4.36e-67 | 37 | 444 | 13 | 449 | Crystalstructure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
| 1FMI_A | 5.06e-67 | 37 | 444 | 18 | 454 | CrystalStructure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
| 1FO2_A | 5.32e-67 | 37 | 444 | 18 | 454 | CrystalStructure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
| 5KK7_A | 2.55e-66 | 37 | 444 | 13 | 449 | Crystalstructure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9C512 | 2.17e-81 | 27 | 445 | 95 | 535 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 OS=Arabidopsis thaliana OX=3702 GN=MNS1 PE=1 SV=1 |
| Q9FG93 | 1.65e-77 | 25 | 444 | 36 | 471 | Alpha-mannosidase I MNS4 OS=Arabidopsis thaliana OX=3702 GN=MNS4 PE=1 SV=1 |
| Q9SXC9 | 7.94e-77 | 25 | 445 | 33 | 477 | Alpha-mannosidase I MNS5 OS=Arabidopsis thaliana OX=3702 GN=MNS5 PE=1 SV=1 |
| Q8H116 | 2.23e-75 | 32 | 445 | 101 | 536 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS2 OS=Arabidopsis thaliana OX=3702 GN=MNS2 PE=1 SV=1 |
| Q9BV94 | 8.85e-71 | 22 | 444 | 28 | 480 | ER degradation-enhancing alpha-mannosidase-like protein 2 OS=Homo sapiens OX=9606 GN=EDEM2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
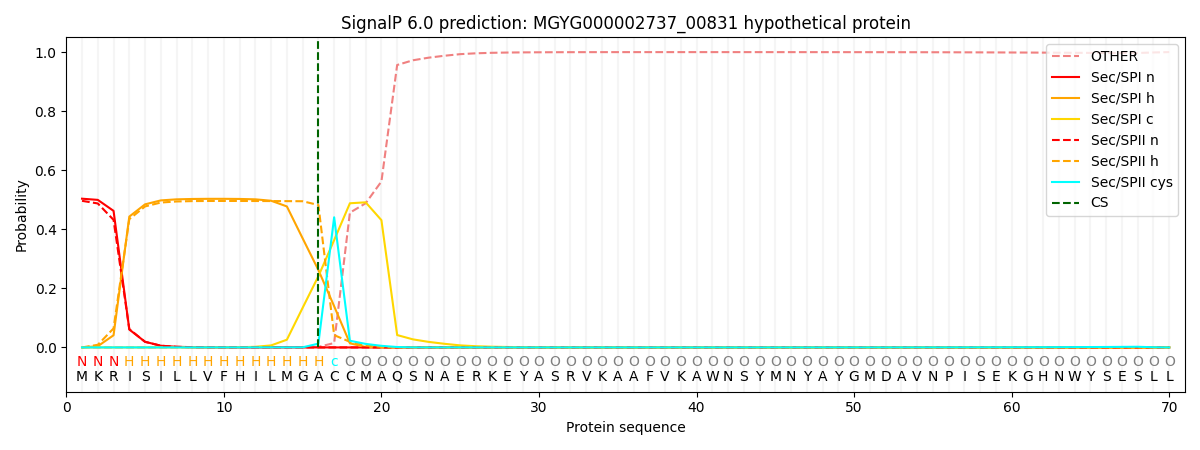
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000676 | 0.492800 | 0.505717 | 0.000286 | 0.000282 | 0.000226 |
