You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002832_01905
You are here: Home > Sequence: MGYG000002832_01905
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900548195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900548195 | |||||||||||
| CAZyme ID | MGYG000002832_01905 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1977; End: 4289 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 457 | 714 | 1.4e-48 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.78e-34 | 459 | 713 | 56 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 4.26e-33 | 459 | 714 | 98 | 309 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.68e-18 | 464 | 677 | 126 | 302 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 4.35e-14 | 49 | 128 | 12 | 90 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 3.24e-08 | 81 | 137 | 1 | 56 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQZ02683.1 | 3.01e-301 | 2 | 770 | 6 | 777 |
| AXH21505.1 | 1.55e-169 | 16 | 719 | 29 | 745 |
| EDV05072.1 | 1.55e-169 | 16 | 719 | 29 | 745 |
| ADE83221.1 | 5.49e-160 | 2 | 769 | 9 | 465 |
| QRX63290.1 | 2.34e-158 | 2 | 719 | 17 | 731 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1W2V_A | 6.23e-17 | 478 | 719 | 118 | 347 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2V_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1CLX_A | 8.27e-17 | 478 | 719 | 117 | 346 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
| 1W2P_A | 8.36e-17 | 478 | 719 | 118 | 347 | The3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2P_B The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W32_A | 8.36e-17 | 478 | 719 | 118 | 347 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W3H_A | 9.44e-17 | 478 | 719 | 129 | 358 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W3H_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14768 | 1.92e-15 | 478 | 719 | 381 | 610 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| O59859 | 2.09e-15 | 448 | 717 | 115 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
| P23360 | 5.44e-14 | 459 | 717 | 128 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q96VB6 | 2.21e-13 | 459 | 717 | 124 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| A2QFV7 | 2.31e-13 | 459 | 717 | 128 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
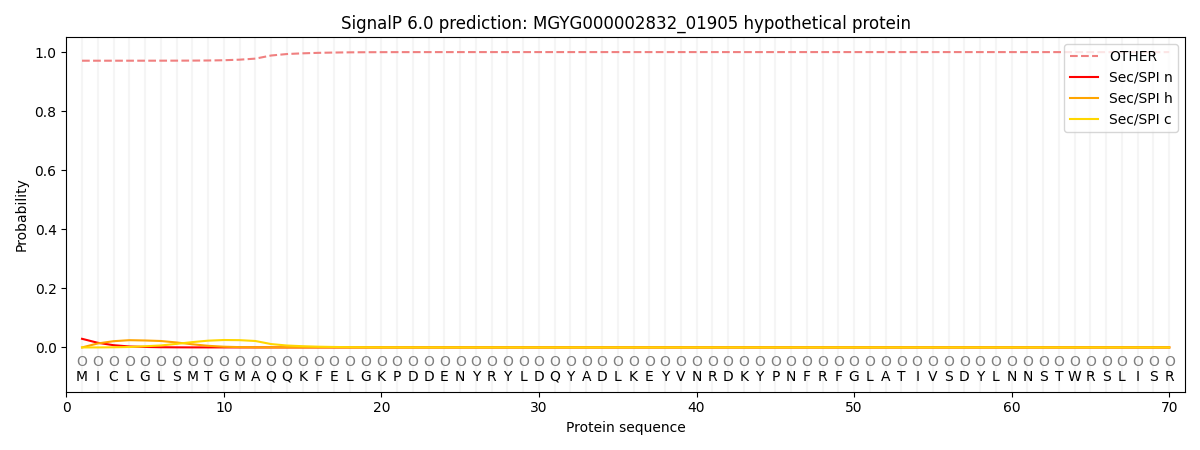
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.973751 | 0.026122 | 0.000059 | 0.000043 | 0.000024 | 0.000038 |
