You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002937_01886
You are here: Home > Sequence: MGYG000002937_01886
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporolactobacillus sp900543345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_G; Sporolactobacillaceae; Sporolactobacillus; Sporolactobacillus sp900543345 | |||||||||||
| CAZyme ID | MGYG000002937_01886 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Cell division suppressor protein YneA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1085; End: 2161 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM50 | 182 | 223 | 1e-16 | 0.95 |
| CBM50 | 315 | 357 | 9e-16 | 0.975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK06347 | PRK06347 | 1.86e-44 | 155 | 354 | 305 | 521 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 1.48e-41 | 141 | 356 | 367 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| COG0791 | Spr | 2.23e-31 | 28 | 151 | 76 | 197 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| pfam00877 | NLPC_P60 | 9.03e-31 | 39 | 152 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK06347 | PRK06347 | 3.73e-25 | 181 | 354 | 264 | 447 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAA26715.1 | 5.82e-110 | 1 | 358 | 1 | 367 |
| QAA23744.1 | 5.82e-110 | 1 | 358 | 1 | 367 |
| BBO00278.1 | 1.17e-109 | 1 | 357 | 1 | 366 |
| QOY35947.1 | 1.42e-49 | 28 | 356 | 30 | 316 |
| ASR40346.1 | 7.44e-43 | 164 | 354 | 639 | 834 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CFL_A | 1.03e-22 | 26 | 123 | 13 | 110 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 3H41_A | 6.36e-11 | 30 | 120 | 192 | 280 | CRYSTALSTRUCTURE OF A NLPC/P60 FAMILY PROTEIN (BCE_2878) FROM BACILLUS CEREUS ATCC 10987 AT 1.79 A RESOLUTION [Bacillus cereus ATCC 10987] |
| 2K1G_A | 4.11e-10 | 37 | 133 | 16 | 108 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 4HPE_A | 1.58e-09 | 28 | 127 | 188 | 287 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 4FDY_A | 2.92e-09 | 28 | 127 | 192 | 291 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5HRU2 | 1.92e-37 | 180 | 358 | 27 | 192 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=sle1 PE=3 SV=1 |
| Q8CMN2 | 1.92e-37 | 180 | 358 | 27 | 192 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=sle1 PE=3 SV=1 |
| Q49UX4 | 2.87e-37 | 180 | 358 | 27 | 195 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| Q7A7E0 | 6.70e-36 | 180 | 358 | 27 | 203 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain N315) OX=158879 GN=sle1 PE=1 SV=1 |
| Q7A1T4 | 6.70e-36 | 180 | 358 | 27 | 203 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain MW2) OX=196620 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
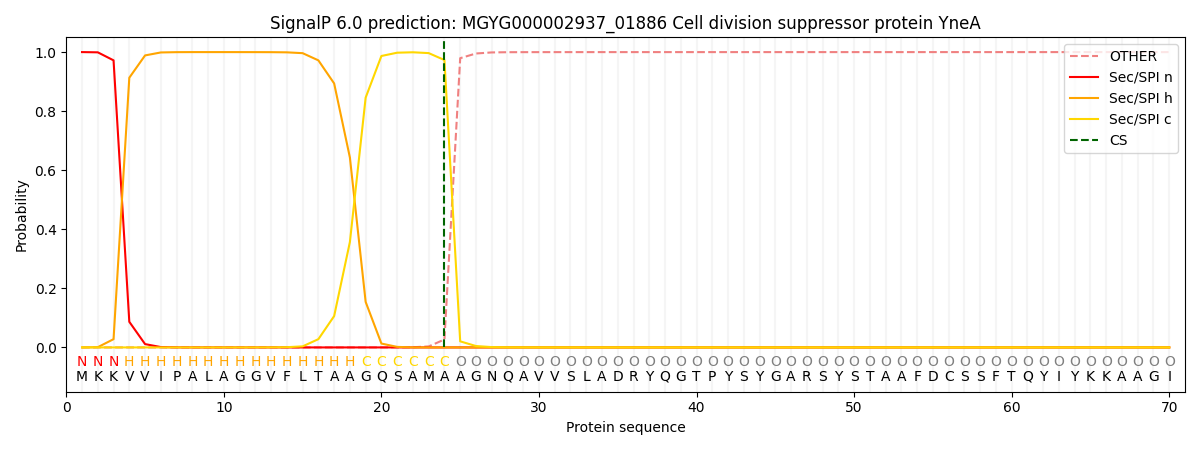
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000259 | 0.999047 | 0.000199 | 0.000194 | 0.000157 | 0.000139 |
