You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003035_02169
You are here: Home > Sequence: MGYG000003035_02169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900542185 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900542185 | |||||||||||
| CAZyme ID | MGYG000003035_02169 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12593; End: 15343 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 21 | 470 | 5.2e-57 | 0.49069148936170215 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.71e-26 | 27 | 456 | 14 | 451 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.01e-10 | 26 | 310 | 13 | 296 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 9.91e-10 | 191 | 290 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK09525 | lacZ | 1.40e-06 | 165 | 311 | 196 | 355 | beta-galactosidase. |
| PRK10340 | ebgA | 4.12e-05 | 165 | 413 | 184 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASM65962.1 | 0.0 | 3 | 916 | 2 | 935 |
| QRP59794.1 | 0.0 | 21 | 916 | 15 | 930 |
| QQT79911.1 | 0.0 | 21 | 916 | 15 | 930 |
| QUU08147.1 | 5.43e-318 | 21 | 916 | 15 | 930 |
| QVJ82312.1 | 4.04e-317 | 26 | 915 | 11 | 877 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ETZ_A | 5.78e-12 | 254 | 433 | 253 | 439 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB [Arthrobacter sp. 32cB],6H1P_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB - data collected at room temperature [Arthrobacter sp. 32cB] |
| 6SEB_A | 5.83e-12 | 254 | 433 | 274 | 460 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG [Arthrobacter sp. 32cB],6SEC_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG [Arthrobacter sp. 32cB],6SED_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose [Arthrobacter sp. 32cB] |
| 6ZJP_A | 5.83e-12 | 254 | 433 | 274 | 460 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q [Arthrobacter sp. 32cB] |
| 6ZJV_A | 5.83e-12 | 254 | 433 | 274 | 460 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
| 6ZJQ_A | 5.84e-12 | 254 | 433 | 276 | 462 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose [Arthrobacter sp. 32cB],6ZJR_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5Z2P7 | 7.27e-11 | 251 | 433 | 289 | 481 | Beta-galactosidase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=lacZ PE=3 SV=1 |
| Q32JB6 | 7.27e-11 | 251 | 433 | 289 | 481 | Beta-galactosidase OS=Shigella dysenteriae serotype 1 (strain Sd197) OX=300267 GN=lacZ PE=3 SV=2 |
| Q8X685 | 7.27e-11 | 251 | 433 | 289 | 481 | Beta-galactosidase OS=Escherichia coli O157:H7 OX=83334 GN=lacZ PE=3 SV=1 |
| B1J0T5 | 3.73e-10 | 251 | 433 | 289 | 481 | Beta-galactosidase OS=Escherichia coli (strain ATCC 8739 / DSM 1576 / NBRC 3972 / NCIMB 8545 / WDCM 00012 / Crooks) OX=481805 GN=lacZ PE=3 SV=1 |
| P00722 | 3.73e-10 | 251 | 433 | 289 | 481 | Beta-galactosidase OS=Escherichia coli (strain K12) OX=83333 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
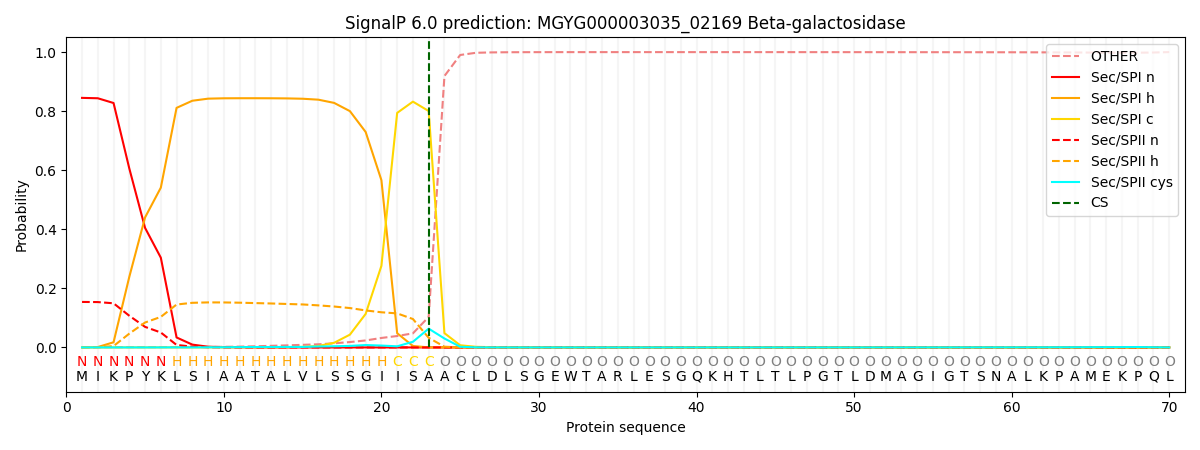
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002692 | 0.835027 | 0.161367 | 0.000355 | 0.000277 | 0.000262 |
