You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003064_00060
You are here: Home > Sequence: MGYG000003064_00060
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides graminisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides graminisolvens | |||||||||||
| CAZyme ID | MGYG000003064_00060 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 88682; End: 91555 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 381 | 878 | 2.9e-101 | 0.5611702127659575 |
| GH105 | 43 | 369 | 2.4e-76 | 0.9819277108433735 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 1.58e-93 | 32 | 370 | 1 | 342 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| PRK10150 | PRK10150 | 2.19e-72 | 433 | 928 | 40 | 576 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 1.24e-70 | 387 | 911 | 14 | 565 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| COG4225 | YesR | 2.89e-38 | 60 | 371 | 44 | 357 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 1.31e-34 | 377 | 884 | 33 | 533 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRX62581.1 | 3.22e-290 | 359 | 951 | 5 | 597 |
| ABQ05112.1 | 3.35e-277 | 371 | 951 | 19 | 599 |
| QLC67640.1 | 6.73e-277 | 358 | 951 | 9 | 599 |
| BCI62566.1 | 6.73e-277 | 357 | 951 | 9 | 600 |
| QSW90199.1 | 9.53e-277 | 371 | 951 | 19 | 599 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D1N_A | 1.11e-243 | 360 | 945 | 11 | 596 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
| 6D89_A | 3.77e-234 | 360 | 945 | 11 | 588 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 7VQM_A | 5.27e-209 | 373 | 951 | 1 | 591 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 5NOA_A | 4.80e-190 | 25 | 370 | 29 | 374 | PolysaccharideLyase BACCELL_00875 [Bacteroides thetaiotaomicron] |
| 6D8K_A | 3.52e-177 | 377 | 950 | 27 | 597 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 6.60e-61 | 404 | 944 | 48 | 628 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| T2KPL9 | 5.76e-49 | 40 | 369 | 44 | 374 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22220 PE=2 SV=1 |
| P05804 | 1.04e-48 | 435 | 941 | 42 | 585 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| L7P9J4 | 1.98e-48 | 40 | 369 | 46 | 375 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Nonlabens ulvanivorans OX=906888 GN=IL45_01505 PE=1 SV=1 |
| O97524 | 4.48e-46 | 361 | 944 | 15 | 625 | Beta-glucuronidase OS=Felis catus OX=9685 GN=GUSB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
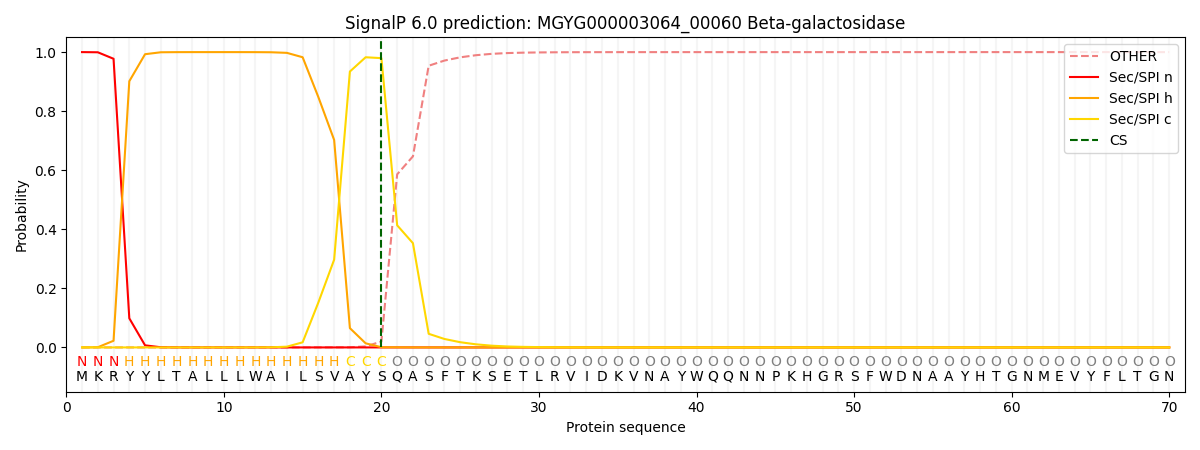
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000411 | 0.998832 | 0.000201 | 0.000192 | 0.000177 | 0.000169 |
