You are browsing environment: HUMAN GUT
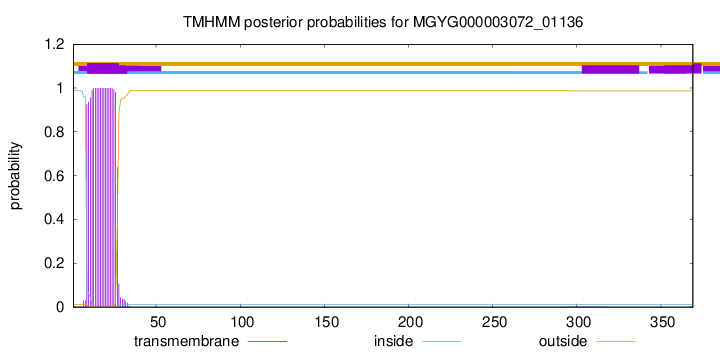
MGYG000003072_01136
Basic Information
help
Species
Paenibacillus amylolyticus
Lineage
Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus
CAZyme ID
MGYG000003072_01136
CAZy Family
GH126
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003072
7042795
MAG
China
Asia
Gene Location
Start: 208172;
End: 209281
Strand: -
No EC number prediction in MGYG000003072_01136.
Family
Start
End
Evalue
family coverage
GH126
40
358
2.6e-74
0.9844236760124611
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam01270
Glyco_hydro_8
1.61e-05
42
176
11
138
Glycosyl hydrolases family 8.
more
COG3405
BcsZ
3.66e-05
76
173
55
156
Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6R2M_A
2.61e-23
35
305
4
265
Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O31486
9.93e-57
38
365
43
357
Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1
more
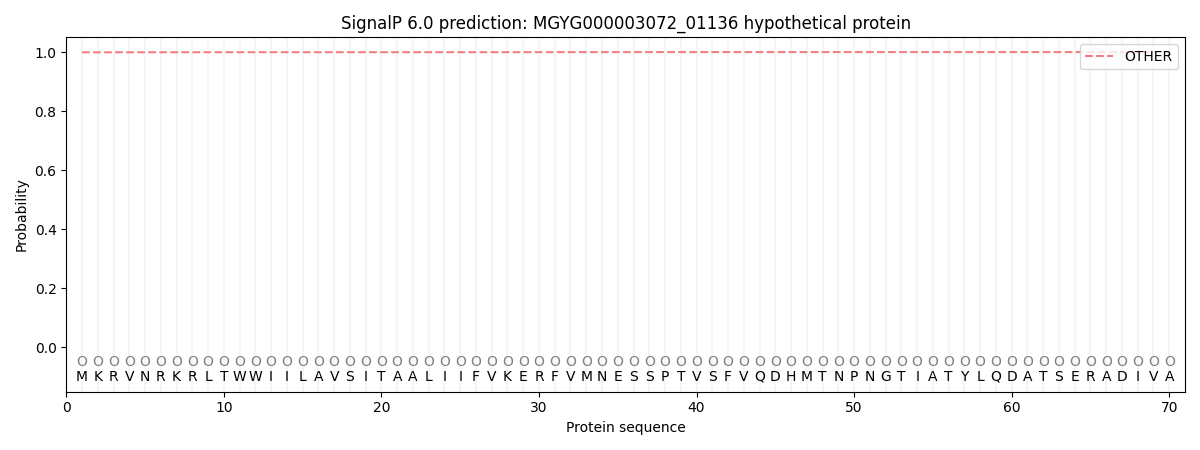
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999354
0.000526
0.000058
0.000004
0.000002
0.000093