You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003115_01864
You are here: Home > Sequence: MGYG000003115_01864
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
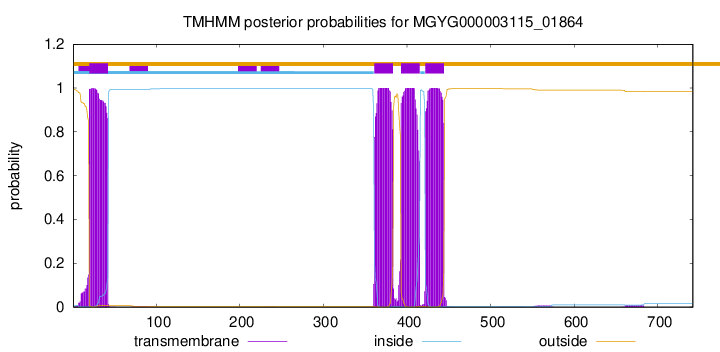
TMHMM annotations
Basic Information help
| Species | Franconibacter helveticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Franconibacter; Franconibacter helveticus | |||||||||||
| CAZyme ID | MGYG000003115_01864 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1685; End: 3913 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 165 | 382 | 2.6e-47 | 0.9847715736040609 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK11234 | nfrB | 0.0 | 8 | 737 | 2 | 727 | phage adsorption protein NrfB. |
| PRK14716 | PRK14716 | 0.0 | 4 | 509 | 4 | 499 | glycosyl transferase family protein. |
| PRK15489 | nfrB | 0.0 | 8 | 714 | 10 | 693 | glycosyl transferase family protein. |
| pfam13632 | Glyco_trans_2_3 | 9.29e-37 | 165 | 383 | 2 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| pfam13641 | Glyco_tranf_2_3 | 1.72e-27 | 71 | 331 | 4 | 228 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJF78978.1 | 0.0 | 1 | 739 | 1 | 739 |
| QLX29339.1 | 0.0 | 1 | 739 | 1 | 739 |
| QLP26252.1 | 0.0 | 1 | 739 | 1 | 739 |
| QLP30795.1 | 0.0 | 1 | 739 | 1 | 739 |
| QDN46533.1 | 0.0 | 1 | 739 | 1 | 739 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AFA5 | 0.0 | 1 | 739 | 1 | 739 | Bacteriophage adsorption protein B OS=Escherichia coli (strain K12) OX=83333 GN=nfrB PE=4 SV=1 |
| P0AFA6 | 0.0 | 1 | 739 | 1 | 739 | Bacteriophage N4 adsorption protein B OS=Escherichia coli O157:H7 OX=83334 GN=nfrB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
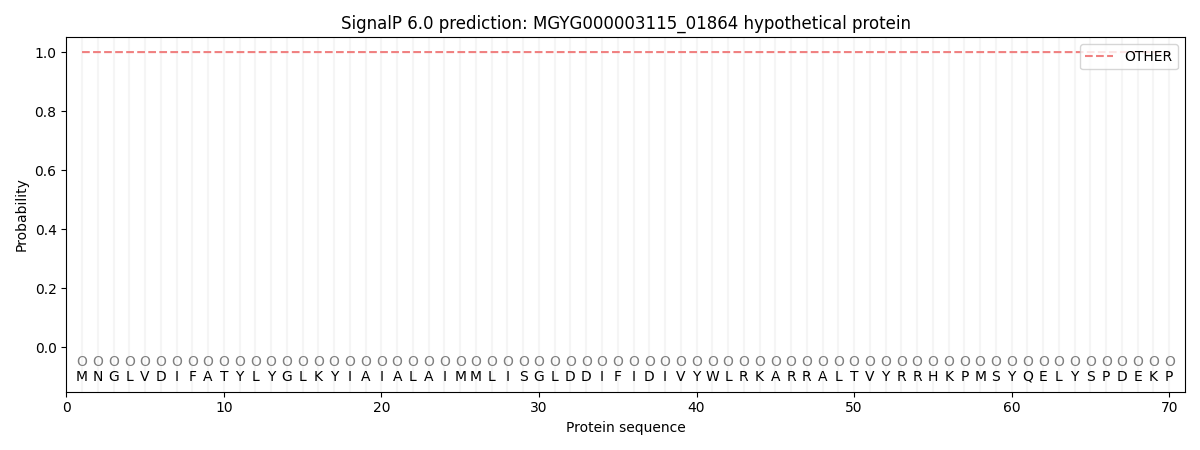
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000065 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |