You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003137_02900
You are here: Home > Sequence: MGYG000003137_02900
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp000015165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp000015165 | |||||||||||
| CAZyme ID | MGYG000003137_02900 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 226307; End: 227863 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5309 | Scw11 | 3.93e-50 | 45 | 281 | 62 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| pfam00332 | Glyco_hydro_17 | 2.01e-05 | 77 | 287 | 59 | 305 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABQ36232.1 | 0.0 | 1 | 518 | 21 | 538 |
| CAL77527.1 | 0.0 | 2 | 518 | 22 | 538 |
| BAM89796.1 | 0.0 | 1 | 518 | 1 | 518 |
| SMX58586.1 | 0.0 | 2 | 518 | 40 | 556 |
| SHH26998.1 | 5.78e-297 | 2 | 516 | 24 | 539 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P43070 | 8.87e-16 | 38 | 287 | 31 | 301 | Glucan 1,3-beta-glucosidase OS=Candida albicans OX=5476 GN=BGL2 PE=3 SV=1 |
| Q5AMT2 | 2.92e-15 | 38 | 287 | 31 | 301 | Glucan 1,3-beta-glucosidase BGL2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BGL2 PE=1 SV=2 |
| P15703 | 4.73e-13 | 34 | 287 | 41 | 306 | Glucan 1,3-beta-glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=BGL2 PE=1 SV=1 |
| P53334 | 6.33e-13 | 20 | 281 | 131 | 386 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
| Q04951 | 6.44e-13 | 25 | 281 | 139 | 389 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999525 | 0.000540 | 0.000002 | 0.000001 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
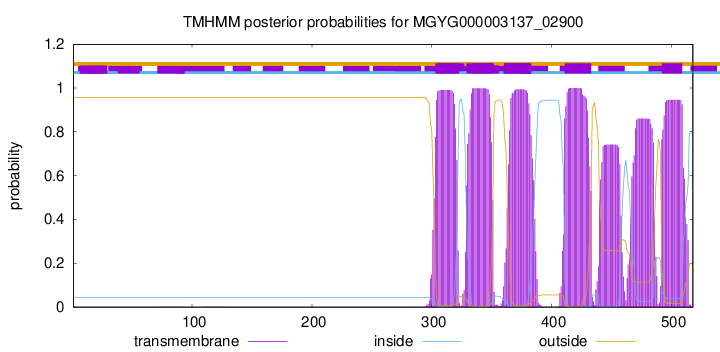
| start | end |
|---|---|
| 303 | 322 |
| 329 | 351 |
| 361 | 383 |
| 411 | 433 |
| 492 | 509 |
