You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003137_05143
Basic Information
help
| Species |
Bradyrhizobium sp000015165
|
| Lineage |
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp000015165
|
| CAZyme ID |
MGYG000003137_05143
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003137 |
8388709 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 57188;
End: 58717
Strand: -
|
No EC number prediction in MGYG000003137_05143.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
12 |
388 |
4.1e-46 |
0.6814814814814815 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13231
|
PMT_2 |
8.80e-39 |
42 |
203 |
1 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1807
|
ArnT |
9.95e-21 |
25 |
364 |
46 |
385 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279
|
arnT |
2.35e-05 |
49 |
199 |
68 |
222 |
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q1MLH5
|
3.03e-17 |
5 |
397 |
9 |
403 |
Lipid A galacturonosyltransferase RgtD OS=Rhizobium leguminosarum bv. viciae (strain 3841) OX=216596 GN=rgtD PE=3 SV=1 |
|
C5BDQ8
|
4.23e-07 |
30 |
359 |
48 |
373 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.816591
|
0.182535
|
0.000241
|
0.000233
|
0.000161
|
0.000234
|
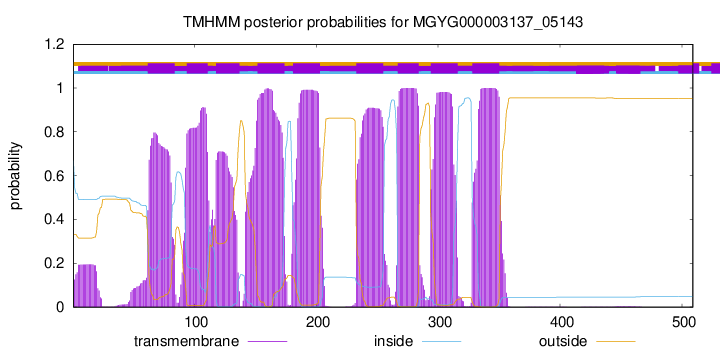
| start |
end |
| 62 |
84 |
| 94 |
111 |
| 118 |
137 |
| 152 |
174 |
| 181 |
203 |
| 233 |
255 |
| 267 |
284 |
| 294 |
316 |
| 328 |
350 |


