You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003172_00085
You are here: Home > Sequence: MGYG000003172_00085
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900555915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900555915 | |||||||||||
| CAZyme ID | MGYG000003172_00085 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5765; End: 8521 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 8.03e-21 | 710 | 859 | 3 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 5.92e-11 | 510 | 563 | 3 | 58 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.08e-08 | 453 | 538 | 1 | 78 | Ricin-type beta-trefoil lectin domain-like. |
| cd00152 | PTX | 8.61e-06 | 725 | 864 | 32 | 176 | Pentraxins are plasma proteins characterized by their pentameric discoid assembly and their Ca2+ dependent ligand binding, such as Serum amyloid P component (SAP) and C-reactive Protein (CRP), which are cytokine-inducible acute-phase proteins implicated in innate immunity. CRP binds to ligands containing phosphocholine, SAP binds to amyloid fibrils, DNA, chromatin, fibronectin, C4-binding proteins and glycosaminoglycans. "Long" pentraxins have N-terminal extensions to the common pentraxin domain; one group, the neuronal pentraxins, may be involved in synapse formation and remodeling, and they may also be able to form heteromultimers. |
| cd00110 | LamG | 9.82e-06 | 705 | 813 | 1 | 108 | Laminin G domain; Laminin G-like domains are usually Ca++ mediated receptors that can have binding sites for steroids, beta1 integrins, heparin, sulfatides, fibulin-1, and alpha-dystroglycans. Proteins that contain LamG domains serve a variety of purposes including signal transduction via cell-surface steroid receptors, adhesion, migration and differentiation through mediation of cell adhesion molecules. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB47762.1 | 1.25e-155 | 38 | 861 | 46 | 837 |
| AGB29926.1 | 8.19e-154 | 40 | 870 | 41 | 861 |
| QIM09902.1 | 2.86e-150 | 54 | 863 | 29 | 824 |
| QUT88791.1 | 2.17e-145 | 37 | 871 | 49 | 853 |
| ALJ60208.1 | 4.27e-145 | 37 | 871 | 49 | 853 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HON_A | 1.58e-06 | 663 | 858 | 1 | 201 | Structureof Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus],5HON_B Structure of Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus] |
| 5HO0_A | 7.84e-06 | 663 | 858 | 640 | 840 | Crystalstructure of AbnA (closed conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HO2_A Crystal structure of AbnA (open conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HOF_A Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose [Geobacillus stearothermophilus],5HP6_A Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state) [Geobacillus stearothermophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
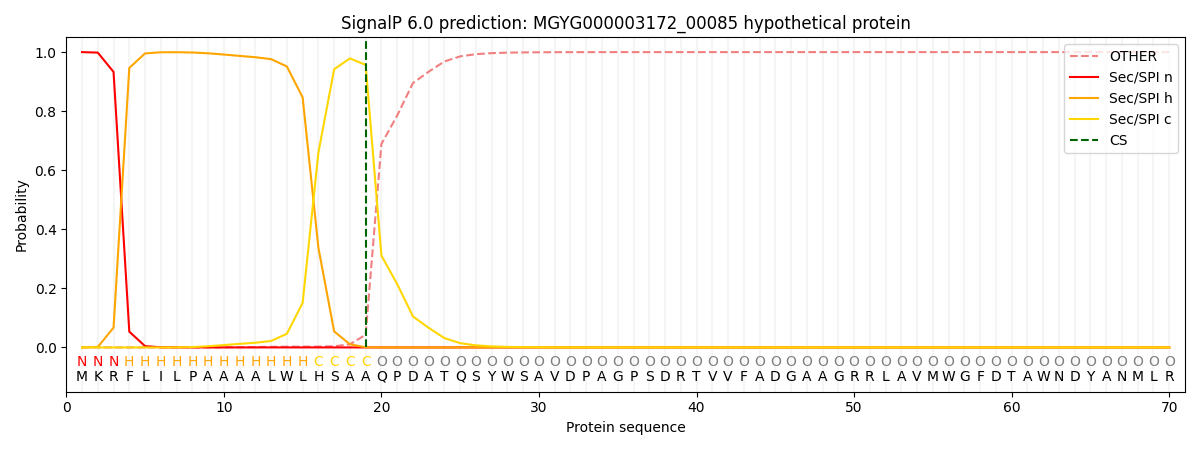
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000851 | 0.998200 | 0.000305 | 0.000230 | 0.000199 | 0.000192 |
