You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003196_00285
You are here: Home > Sequence: MGYG000003196_00285
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
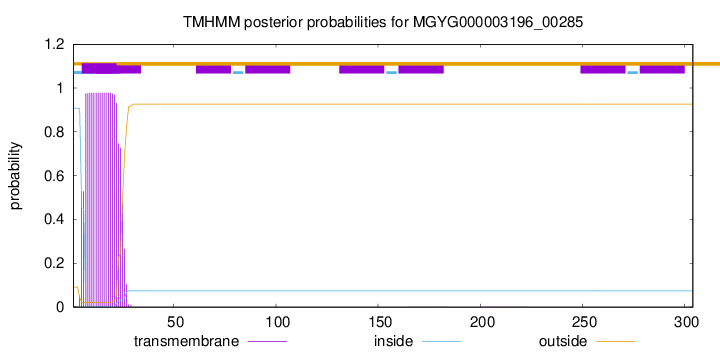
TMHMM annotations
Basic Information help
| Species | Corynebacterium sp900539985 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium sp900539985 | |||||||||||
| CAZyme ID | MGYG000003196_00285 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6686; End: 7600 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 51 | 293 | 2e-32 | 0.9682539682539683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01083 | Cutinase | 5.55e-08 | 51 | 295 | 2 | 171 | Cutinase. |
| pfam08237 | PE-PPE | 3.45e-05 | 104 | 191 | 10 | 94 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHP88711.1 | 1.07e-55 | 43 | 304 | 62 | 329 |
| BCI55762.1 | 2.99e-55 | 43 | 304 | 62 | 329 |
| BBY81420.1 | 2.34e-54 | 42 | 304 | 58 | 329 |
| BBY50964.1 | 6.55e-54 | 43 | 304 | 62 | 329 |
| BBY74844.1 | 1.30e-53 | 51 | 304 | 71 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AJA_A | 8.53e-54 | 43 | 304 | 29 | 296 | CrystalStructure of MSMEG_6394 [Mycolicibacterium smegmatis MC2 155],3AJA_B Crystal Structure of MSMEG_6394 [Mycolicibacterium smegmatis MC2 155] |
| 5W95_A | 2.46e-48 | 46 | 304 | 16 | 279 | ChainA, Conserved membrane protein of uncharacterised function [Mycobacterium tuberculosis],5W95_B Chain B, Conserved membrane protein of uncharacterised function [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8NLR5 | 1.61e-124 | 1 | 302 | 1 | 307 | Carboxylesterase Culp6 homolog OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=Cgl2873 PE=1 SV=1 |
| A0R619 | 1.19e-52 | 43 | 304 | 64 | 331 | Carboxylesterase Culp6 homolog OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=MSMEG_6394 PE=1 SV=1 |
| O53581 | 2.73e-46 | 51 | 304 | 72 | 330 | Carboxylesterase/lipase Culp6 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut6 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.529980 | 0.460179 | 0.007425 | 0.000861 | 0.000461 | 0.001079 |