You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00769
You are here: Home > Sequence: MGYG000003205_00769
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00769 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33537; End: 34955 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 55 | 453 | 2.9e-144 | 0.9899749373433584 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 7.36e-15 | 58 | 438 | 3 | 335 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 2.86e-08 | 59 | 184 | 1 | 117 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 5.61e-07 | 57 | 211 | 3 | 146 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV99796.1 | 7.74e-173 | 3 | 471 | 5 | 480 |
| AWO02107.1 | 1.45e-164 | 3 | 468 | 5 | 469 |
| BAX78835.1 | 9.34e-162 | 3 | 469 | 5 | 478 |
| QMU30554.1 | 1.92e-160 | 3 | 469 | 5 | 467 |
| AHW60523.1 | 1.23e-159 | 3 | 469 | 5 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 8.16e-83 | 48 | 458 | 32 | 444 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 7.80e-67 | 46 | 453 | 11 | 432 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 3NTO_A | 7.72e-09 | 59 | 390 | 3 | 295 | Crystalstructure of K97V mutant myo-inositol dehydrogenase from Bacillus subtilis [Bacillus subtilis],3NTQ_A Crystal structure of K97V mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD [Bacillus subtilis],3NTQ_B Crystal structure of K97V mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD [Bacillus subtilis],3NTR_A Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol [Bacillus subtilis],3NTR_B Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8H2K3 | 2.47e-110 | 1 | 460 | 3 | 453 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
| Q0HKG4 | 8.61e-109 | 1 | 460 | 4 | 456 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_1375 PE=3 SV=1 |
| Q0HWR6 | 1.72e-108 | 1 | 460 | 4 | 456 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain MR-7) OX=60481 GN=Shewmr7_1440 PE=3 SV=1 |
| A0KV43 | 2.43e-108 | 1 | 460 | 4 | 456 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_1428 PE=3 SV=1 |
| Q8ECL7 | 2.73e-107 | 1 | 460 | 4 | 456 | Alpha-N-acetylgalactosaminidase OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=nagA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
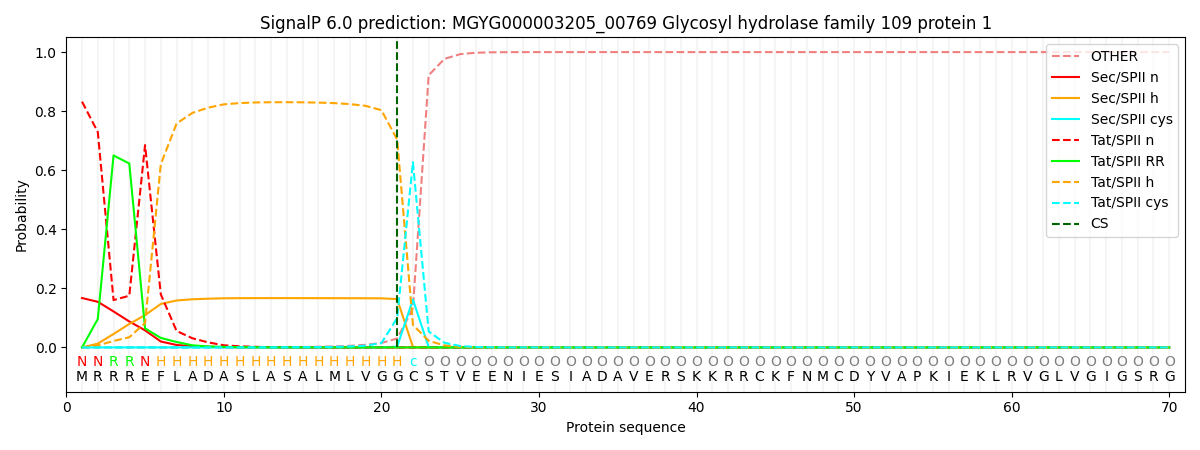
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000005 | 0.000006 | 0.167260 | 0.000487 | 0.832258 | 0.000000 |
