You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00862
You are here: Home > Sequence: MGYG000003205_00862
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00862 | |||||||||||
| CAZy Family | GH42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 153237; End: 155888 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 222 | 476 | 2.8e-17 | 0.6684636118598383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 6.33e-16 | 225 | 446 | 20 | 231 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 1.13e-05 | 222 | 475 | 37 | 285 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV74990.1 | 1.62e-131 | 30 | 872 | 32 | 873 |
| SDT33465.1 | 3.68e-56 | 249 | 823 | 127 | 718 |
| QHI69172.1 | 4.45e-42 | 223 | 854 | 276 | 924 |
| QIK60332.1 | 1.99e-12 | 225 | 523 | 57 | 331 |
| QIK54901.1 | 3.12e-11 | 225 | 523 | 57 | 331 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UCF_A | 5.28e-06 | 223 | 431 | 42 | 242 | Crystalstructure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_B Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_C Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UZS_A Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_B Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_C Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
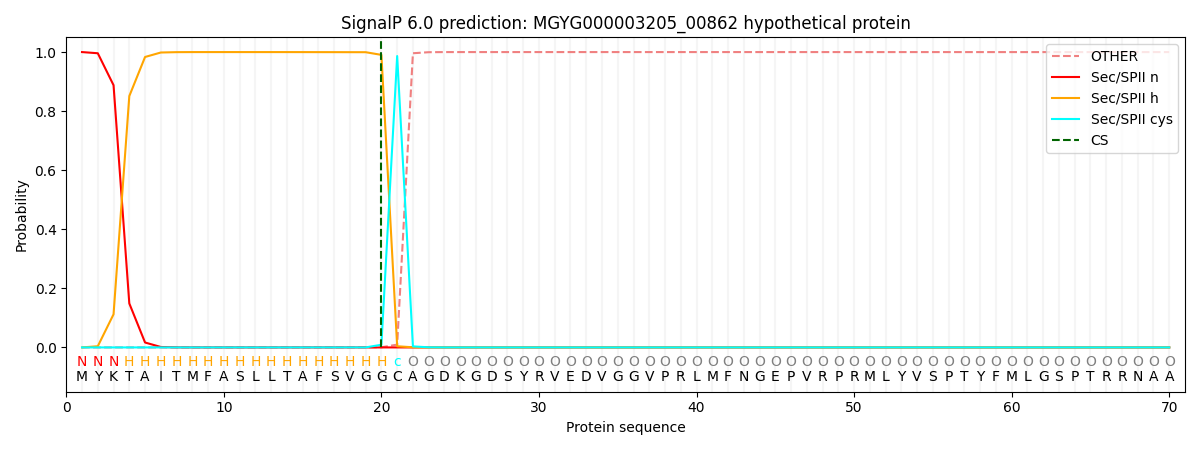
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000062 | 0.000000 | 0.000000 | 0.000000 |
