You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003219_00555
Basic Information
help
| Species |
HGM12657 sp900760575
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; HGM12657; HGM12657 sp900760575
|
| CAZyme ID |
MGYG000003219_00555
|
| CAZy Family |
GT84 |
| CAZyme Description |
Cyclic beta-(1,2)-glucan synthase NdvB
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 549 |
|
60682.14 |
8.8051 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003219 |
1949652 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 3;
End: 1652
Strand: +
|
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam11329
|
DUF3131 |
0.002 |
371 |
479 |
1 |
94 |
Protein of unknown function (DUF3131). This bacterial family of proteins has no known function. |
| COG2943
|
MdoH |
0.008 |
110 |
280 |
347 |
536 |
Membrane glycosyltransferase [Cell wall/membrane/envelope biogenesis, Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P20471
|
1.59e-70 |
1 |
487 |
625 |
1117 |
Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000063
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
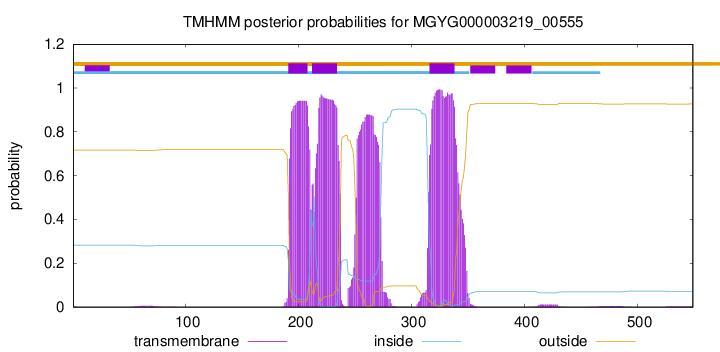
| start |
end |
| 191 |
208 |
| 212 |
234 |
| 316 |
338 |


