You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003237_02059
You are here: Home > Sequence: MGYG000003237_02059
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp002998355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355 | |||||||||||
| CAZyme ID | MGYG000003237_02059 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53117; End: 55330 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 399 | 733 | 4.1e-72 | 0.9678362573099415 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 3.86e-79 | 396 | 733 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13385 | Laminin_G_3 | 1.09e-09 | 60 | 206 | 15 | 149 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam13088 | BNR_2 | 2.02e-05 | 412 | 714 | 54 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| smart00560 | LamGL | 3.20e-05 | 63 | 205 | 2 | 133 | LamG-like jellyroll fold domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44867.1 | 0.0 | 1 | 737 | 1 | 737 |
| QEC45250.1 | 1.08e-52 | 388 | 729 | 88 | 430 |
| VIP09812.1 | 2.90e-52 | 397 | 734 | 26 | 357 |
| QJB38038.1 | 9.22e-51 | 390 | 734 | 20 | 376 |
| QDU26291.1 | 2.46e-50 | 378 | 717 | 9 | 339 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EUR_A | 3.24e-28 | 402 | 719 | 18 | 346 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 1W8N_A | 5.52e-28 | 402 | 719 | 14 | 342 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1EUT_A | 4.18e-27 | 402 | 719 | 18 | 346 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1WCQ_A | 1.28e-26 | 402 | 719 | 14 | 342 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BZD_A | 2.26e-26 | 402 | 719 | 14 | 342 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 2.79e-26 | 402 | 719 | 60 | 388 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| A5PF10 | 1.16e-23 | 409 | 720 | 78 | 398 | Sialidase-1 OS=Sus scrofa OX=9823 GN=NEU1 PE=3 SV=1 |
| Q99519 | 3.70e-23 | 409 | 720 | 77 | 397 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
| Q5RAF4 | 3.70e-23 | 409 | 720 | 77 | 397 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
| P29767 | 4.42e-23 | 372 | 732 | 358 | 821 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
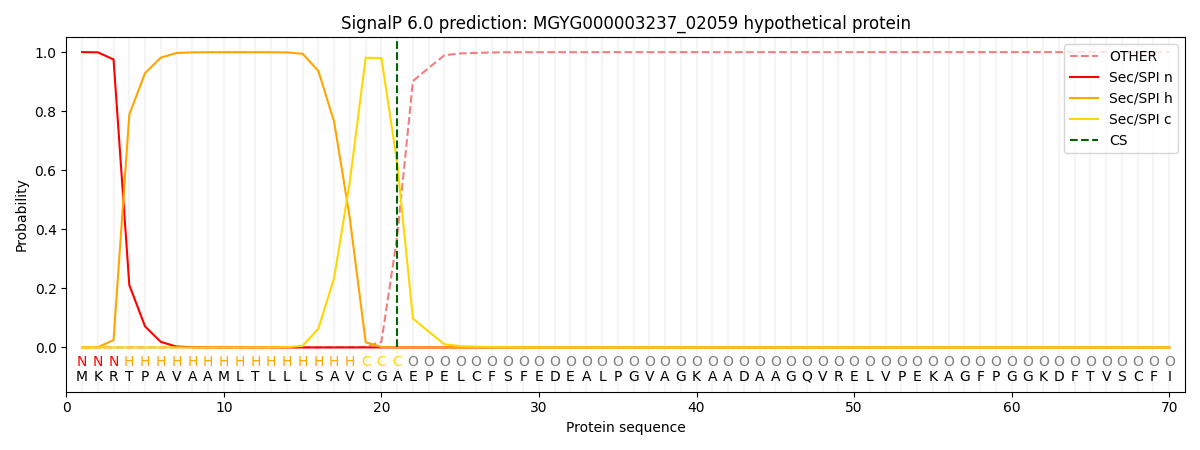
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000552 | 0.998633 | 0.000227 | 0.000207 | 0.000171 | 0.000162 |
