You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003245_00856
You are here: Home > Sequence: MGYG000003245_00856
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
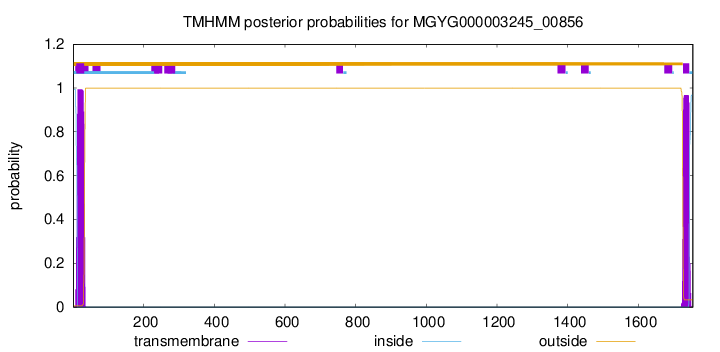
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900761655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900761655 | |||||||||||
| CAZyme ID | MGYG000003245_00856 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7735; End: 13002 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 254 | 383 | 5.3e-17 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14244 | GH_101_like | 2.45e-50 | 570 | 882 | 1 | 297 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam08305 | NPCBM | 2.58e-17 | 251 | 384 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 6.47e-17 | 47 | 244 | 3 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.79e-16 | 1200 | 1395 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 8.52e-13 | 118 | 243 | 47 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD87425.1 | 8.66e-156 | 119 | 1001 | 76 | 1042 |
| QTH41000.1 | 7.92e-126 | 358 | 1048 | 135 | 821 |
| SYX82446.1 | 1.09e-122 | 449 | 1057 | 16 | 632 |
| SMF83630.1 | 9.70e-117 | 491 | 1076 | 57 | 648 |
| AZS16787.1 | 2.82e-95 | 1 | 794 | 1 | 691 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VMG_A | 1.02e-06 | 1191 | 1375 | 5 | 143 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMH_A | 2.28e-06 | 1194 | 1375 | 2 | 137 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMI_A | 2.28e-06 | 1194 | 1375 | 2 | 137 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 7.04e-13 | 1477 | 1673 | 1667 | 1872 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| P36905 | 5.26e-06 | 1070 | 1190 | 896 | 1011 | Amylopullulanase OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=apu PE=3 SV=2 |
| P38536 | 9.60e-06 | 1112 | 1185 | 929 | 1004 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
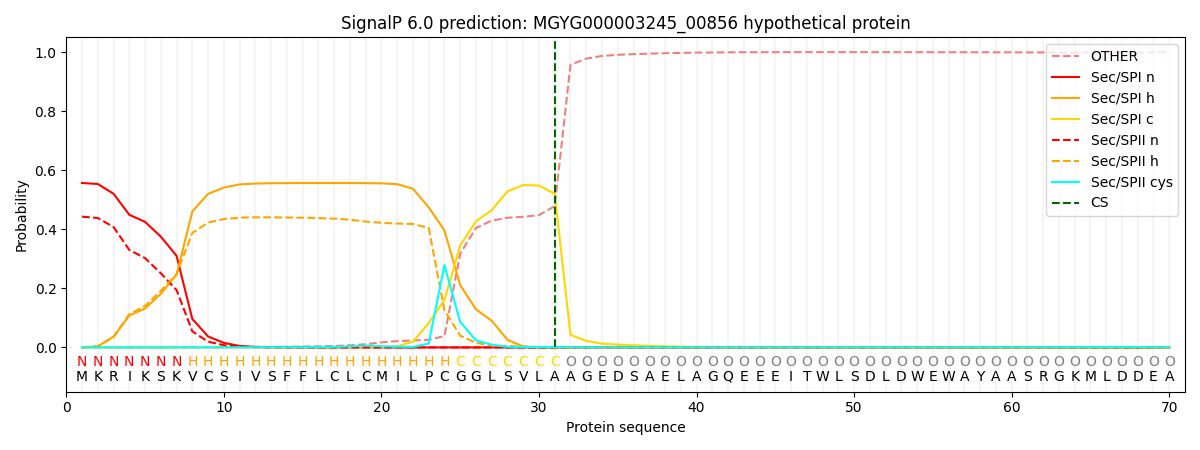
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001144 | 0.548635 | 0.449514 | 0.000275 | 0.000215 | 0.000196 |