You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003250_01346
You are here: Home > Sequence: MGYG000003250_01346
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
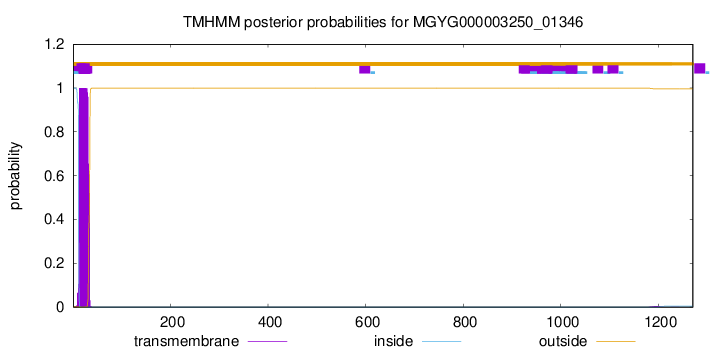
TMHMM annotations
Basic Information help
| Species | UMGS1880 sp900761765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-552; UMGS1880; UMGS1880 sp900761765 | |||||||||||
| CAZyme ID | MGYG000003250_01346 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5013; End: 8828 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 731 | 905 | 2.7e-34 | 0.7870370370370371 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01915 | Glyco_hydro_3_C | 2.37e-14 | 131 | 332 | 2 | 184 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 5.37e-14 | 97 | 510 | 351 | 737 | beta-glucosidase BglX. |
| pfam01391 | Collagen | 6.08e-12 | 33 | 68 | 18 | 53 | Collagen triple helix repeat (20 copies). Members of this family belong to the collagen superfamily. Collagens are generally extracellular structural proteins involved in formation of connective tissue structure. The alignment contains 20 copies of the G-X-Y repeat that forms a triple helix. The first position of the repeat is glycine, the second and third positions can be any residue but are frequently proline and hydroxy-proline. Collagens are post translationally modified by proline hydroxylase to form the hydroxy-proline residues. Defective hydroxylation is the cause of scurvy. Some members of the collagen superfamily are not involved in connective tissue structure but share the same triple helical structure. The family includes bacterial collagen-like triple-helix repeat proteins. |
| pfam00933 | Glyco_hydro_3 | 6.64e-12 | 732 | 901 | 84 | 255 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01391 | Collagen | 3.09e-10 | 33 | 73 | 6 | 46 | Collagen triple helix repeat (20 copies). Members of this family belong to the collagen superfamily. Collagens are generally extracellular structural proteins involved in formation of connective tissue structure. The alignment contains 20 copies of the G-X-Y repeat that forms a triple helix. The first position of the repeat is glycine, the second and third positions can be any residue but are frequently proline and hydroxy-proline. Collagens are post translationally modified by proline hydroxylase to form the hydroxy-proline residues. Defective hydroxylation is the cause of scurvy. Some members of the collagen superfamily are not involved in connective tissue structure but share the same triple helical structure. The family includes bacterial collagen-like triple-helix repeat proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEU80230.1 | 2.00e-158 | 103 | 1092 | 28 | 1009 |
| QOS39237.1 | 4.78e-144 | 100 | 1078 | 43 | 1034 |
| VEU80232.1 | 4.56e-140 | 104 | 924 | 66 | 882 |
| QEN05281.1 | 3.52e-126 | 73 | 922 | 37 | 883 |
| QOL35078.1 | 5.52e-125 | 87 | 922 | 51 | 874 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 4.34e-58 | 126 | 920 | 44 | 763 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X42_A | 2.24e-21 | 690 | 917 | 3 | 258 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 2X40_A | 2.24e-21 | 690 | 917 | 3 | 258 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 3AC0_A | 8.95e-17 | 692 | 917 | 7 | 241 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 5NBS_A | 2.70e-16 | 121 | 429 | 387 | 656 | Structuralstudies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa [Neurospora crassa OR74A],5NBS_B Structural studies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa [Neurospora crassa OR74A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15885 | 3.78e-54 | 122 | 903 | 13 | 699 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| P16084 | 4.32e-53 | 113 | 902 | 20 | 771 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| A1CA51 | 1.96e-20 | 692 | 992 | 7 | 310 | Probable beta-glucosidase I OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=bglI PE=3 SV=1 |
| P27034 | 7.52e-20 | 692 | 917 | 3 | 238 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q2U8Y5 | 7.73e-20 | 692 | 917 | 7 | 241 | Probable beta-glucosidase I OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
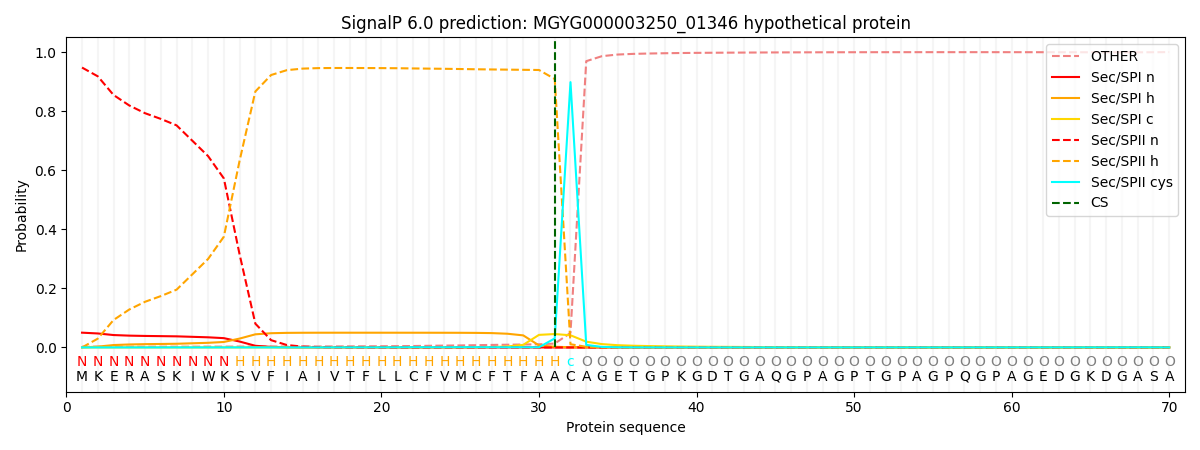
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002864 | 0.047680 | 0.949196 | 0.000069 | 0.000093 | 0.000073 |