You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003312_01035
You are here: Home > Sequence: MGYG000003312_01035
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900547205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900547205 | |||||||||||
| CAZyme ID | MGYG000003312_01035 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4273; End: 5598 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 310 | 436 | 8.1e-19 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08522 | DUF1735 | 1.19e-12 | 44 | 143 | 18 | 115 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 2.89e-12 | 319 | 436 | 16 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam08522 | DUF1735 | 5.96e-11 | 165 | 266 | 7 | 115 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU08575.1 | 3.22e-277 | 1 | 441 | 1 | 441 |
| ASM66238.1 | 3.22e-277 | 1 | 441 | 1 | 441 |
| QQT77186.1 | 3.22e-277 | 1 | 441 | 1 | 441 |
| QRP57568.1 | 3.22e-277 | 1 | 441 | 1 | 441 |
| QMW86943.1 | 1.45e-225 | 1 | 440 | 1 | 439 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GGL_A | 3.86e-29 | 284 | 440 | 1 | 157 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 3F2Z_A | 3.99e-28 | 295 | 440 | 2 | 147 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 2KD7_A | 5.21e-26 | 295 | 440 | 2 | 147 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
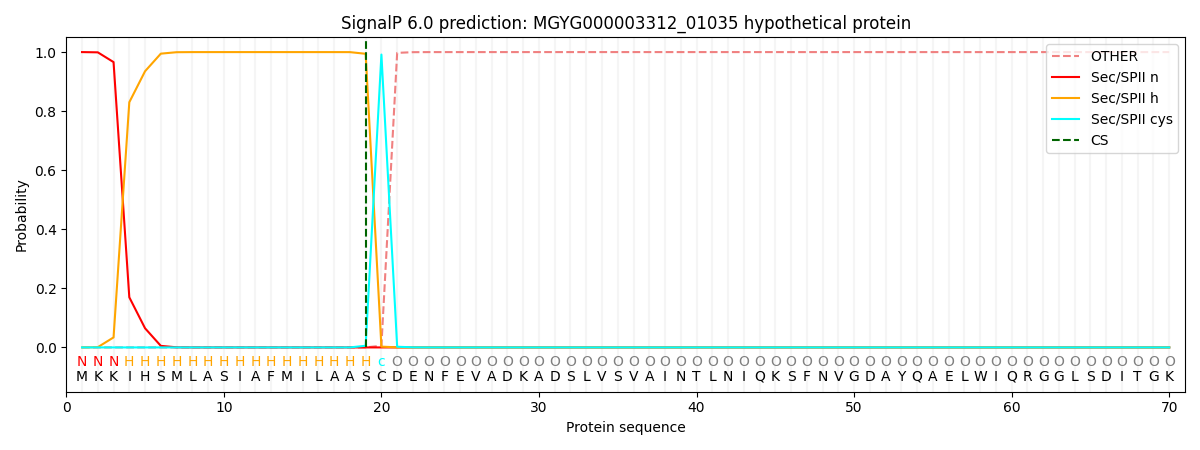
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000035 | 0.000000 | 0.000000 | 0.000000 |
