You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003375_00541
You are here: Home > Sequence: MGYG000003375_00541
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium striatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium striatum | |||||||||||
| CAZyme ID | MGYG000003375_00541 | |||||||||||
| CAZy Family | GT53 | |||||||||||
| CAZyme Description | putative arabinosyltransferase C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 112246; End: 115611 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT53 | 20 | 1112 | 0 | 0.9971401334604385 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04602 | Arabinose_trans | 0.0 | 206 | 650 | 1 | 445 | Mycobacterial cell wall arabinan synthesis protein. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. |
| pfam14896 | Arabino_trans_C | 2.34e-178 | 713 | 1112 | 1 | 384 | EmbC C-terminal domain. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. This domain represents the C-terminal extracellular domain that is likely to bind to carbohydrate. |
| pfam17689 | Arabino_trans_N | 1.21e-50 | 46 | 202 | 1 | 155 | Arabinosyltransferase concanavalin like domain. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQE52411.1 | 0.0 | 1 | 1121 | 1 | 1121 |
| ATZ09923.1 | 0.0 | 8 | 1121 | 1 | 1114 |
| ATZ07310.1 | 0.0 | 8 | 1121 | 1 | 1114 |
| ART20129.1 | 0.0 | 1 | 1121 | 1 | 1121 |
| AMO88994.1 | 0.0 | 1 | 1121 | 1 | 1121 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7BVE_A | 1.08e-259 | 20 | 1113 | 13 | 1073 | Cryo-EMstructure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155],7BVE_B Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155] |
| 7BVH_A | 2.67e-259 | 20 | 1113 | 26 | 1086 | Crystalstructure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose [Mycolicibacterium smegmatis MC2 155],7BVH_B Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose [Mycolicibacterium smegmatis MC2 155] |
| 7BWR_A | 4.26e-236 | 20 | 1115 | 24 | 1082 | Mycobacteriumsmegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric 'active state' [Mycolicibacterium smegmatis MC2 155],7BWR_B Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric 'active state' [Mycolicibacterium smegmatis MC2 155],7BX8_A Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric 'resting state' [Mycolicibacterium smegmatis MC2 155],7BX8_B Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric 'resting state' [Mycolicibacterium smegmatis MC2 155] |
| 6X0O_A | 6.89e-236 | 20 | 1115 | 24 | 1082 | Single-ParticleCryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis [Mycolicibacterium smegmatis MC2 155] |
| 7BVC_B | 7.31e-236 | 20 | 1115 | 24 | 1082 | Cryo-EMstructure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155],7BVG_B Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. [Mycolicibacterium smegmatis MC2 155] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q50393 | 3.04e-256 | 20 | 1113 | 13 | 1073 | Probable arabinosyltransferase C OS=Mycolicibacterium smegmatis OX=1772 GN=embC PE=3 SV=2 |
| Q7TVN4 | 4.55e-242 | 20 | 1106 | 27 | 1086 | Probable arabinosyltransferase C OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=embC PE=3 SV=1 |
| Q9CDA7 | 2.44e-241 | 23 | 1106 | 12 | 1063 | Probable arabinosyltransferase C OS=Mycobacterium leprae (strain TN) OX=272631 GN=embC PE=3 SV=1 |
| P9WNL5 | 2.54e-241 | 20 | 1106 | 27 | 1086 | Probable arabinosyltransferase C OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=embC PE=1 SV=1 |
| P9WNL4 | 3.58e-241 | 20 | 1106 | 27 | 1086 | Probable arabinosyltransferase C OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=embC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000003 | 0.000008 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
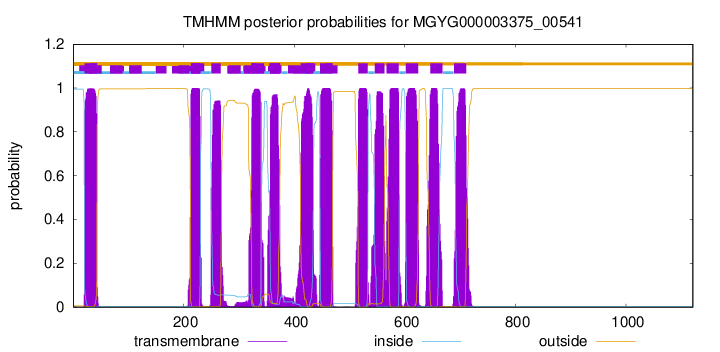
| start | end |
|---|---|
| 21 | 43 |
| 213 | 230 |
| 250 | 267 |
| 323 | 345 |
| 357 | 374 |
| 412 | 434 |
| 447 | 469 |
| 516 | 533 |
| 546 | 563 |
| 567 | 589 |
| 602 | 624 |
| 646 | 668 |
| 689 | 711 |
