You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003423_01788
You are here: Home > Sequence: MGYG000003423_01788
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | W3P20-009 sp900766825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; W3P20-009; W3P20-009; W3P20-009 sp900766825 | |||||||||||
| CAZyme ID | MGYG000003423_01788 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7912; End: 10902 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 22 | 720 | 4.2e-61 | 0.6795212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.90e-31 | 18 | 481 | 1 | 434 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.15e-12 | 79 | 452 | 69 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.05e-11 | 278 | 475 | 274 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 1.71e-09 | 273 | 454 | 283 | 464 | beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 2.81e-08 | 323 | 475 | 1 | 157 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY36269.1 | 5.26e-274 | 32 | 990 | 27 | 927 |
| QUT92878.1 | 1.06e-269 | 1 | 992 | 1 | 936 |
| ALJ61552.1 | 5.99e-269 | 1 | 992 | 1 | 936 |
| QUT68948.1 | 5.43e-267 | 17 | 992 | 11 | 936 |
| QUU00164.1 | 7.68e-267 | 17 | 992 | 11 | 936 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6CVM_A | 1.34e-08 | 106 | 454 | 151 | 462 | Atomicresolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_B Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_C Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_D Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6X1Q_A 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_B 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_C 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_D 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12] |
| 3I3B_A | 1.76e-08 | 106 | 454 | 152 | 463 | ChainA, Beta-galactosidase [Escherichia coli K-12],3I3B_B Chain B, Beta-galactosidase [Escherichia coli K-12],3I3B_C Chain C, Beta-galactosidase [Escherichia coli K-12],3I3B_D Chain D, Beta-galactosidase [Escherichia coli K-12],3I3D_A Chain A, Beta-galactosidase [Escherichia coli K-12],3I3D_B Chain B, Beta-galactosidase [Escherichia coli K-12],3I3D_C Chain C, Beta-galactosidase [Escherichia coli K-12],3I3D_D Chain D, Beta-galactosidase [Escherichia coli K-12],3I3E_A Chain A, Beta-galactosidase [Escherichia coli K-12],3I3E_B Chain B, Beta-galactosidase [Escherichia coli K-12],3I3E_C Chain C, Beta-galactosidase [Escherichia coli K-12],3I3E_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
| 1F4A_A | 1.76e-08 | 106 | 454 | 150 | 461 | E.COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_B E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_C E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_D E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4H_A E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_B E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_C E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_D E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli] |
| 3IAP_A | 1.76e-08 | 106 | 454 | 152 | 463 | ChainA, Beta-galactosidase [Escherichia coli K-12],3IAP_B Chain B, Beta-galactosidase [Escherichia coli K-12],3IAP_C Chain C, Beta-galactosidase [Escherichia coli K-12],3IAP_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
| 3MUY_1 | 1.76e-08 | 106 | 454 | 152 | 463 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7MN76 | 8.26e-11 | 106 | 458 | 163 | 477 | Beta-galactosidase OS=Cronobacter sakazakii (strain ATCC BAA-894) OX=290339 GN=lacZ PE=3 SV=1 |
| A7KGA5 | 1.64e-09 | 106 | 454 | 153 | 464 | Beta-galactosidase OS=Klebsiella pneumoniae OX=573 GN=lacZ PE=3 SV=1 |
| A6TI29 | 1.64e-09 | 106 | 454 | 153 | 464 | Beta-galactosidase 2 OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=lacZ2 PE=3 SV=1 |
| B1J0T5 | 3.26e-08 | 106 | 454 | 153 | 464 | Beta-galactosidase OS=Escherichia coli (strain ATCC 8739 / DSM 1576 / NBRC 3972 / NCIMB 8545 / WDCM 00012 / Crooks) OX=481805 GN=lacZ PE=3 SV=1 |
| A7ZWZ1 | 4.28e-08 | 106 | 454 | 153 | 464 | Beta-galactosidase OS=Escherichia coli O9:H4 (strain HS) OX=331112 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
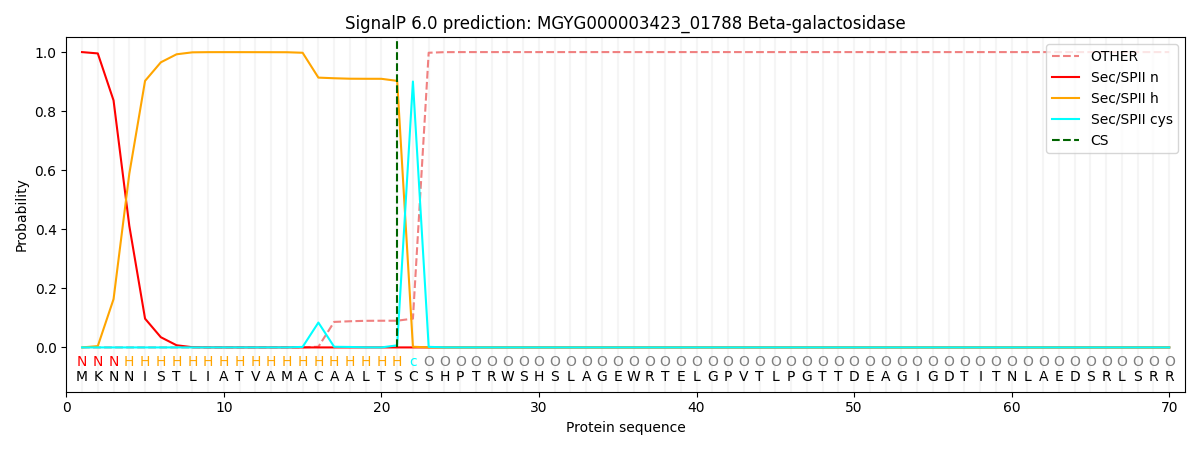
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000020 | 0.000000 | 0.000000 | 0.000000 |
