You are browsing environment: HUMAN GUT
MGYG000003449_01696
Basic Information
help
Species
CAG-312 sp002405555
Lineage
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp002405555
CAZyme ID
MGYG000003449_01696
CAZy Family
GH106
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003449
2395522
MAG
Fiji
Oceania
Gene Location
Start: 151983;
End: 155222
Strand: +
No EC number prediction in MGYG000003449_01696.
Family
Start
End
Evalue
family coverage
GH106
54
849
1.2e-59
0.9368932038834952
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam17132
Glyco_hydro_106
3.42e-13
202
585
401
760
alpha-L-rhamnosidase.
more
pfam14324
PINIT
1.27e-04
793
850
48
119
PINIT domain. The PINIT domain is a protein domain that is found in PIAS proteins. The PINIT domain is about 180 amino acids in length.
more
pfam08531
Bac_rhamnosid_N
0.002
809
888
17
108
Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
T2KNA8
5.62e-09
202
1021
264
1098
Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1
more
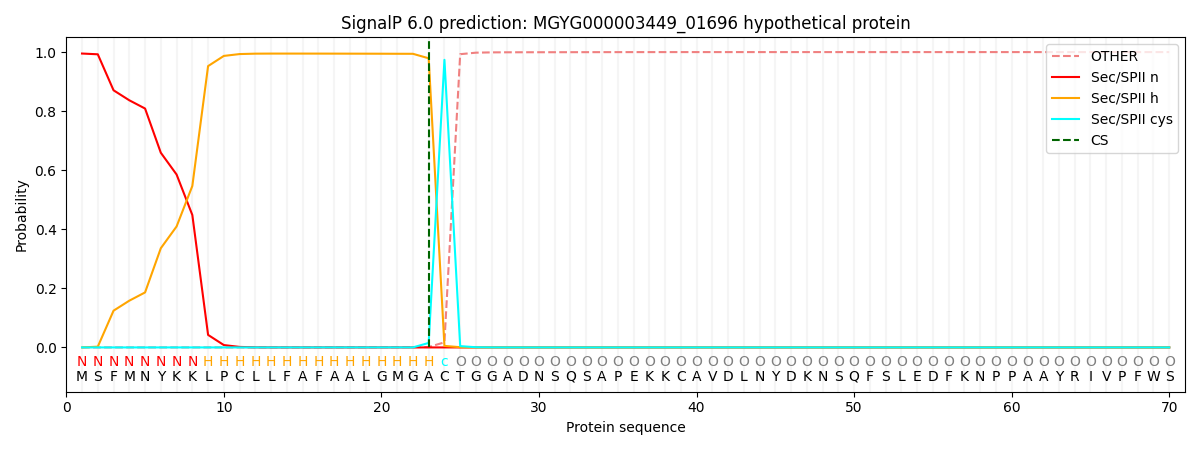
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000019
0.004663
0.995345
0.000002
0.000004
0.000003
There is no transmembrane helices in MGYG000003449_01696.