You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003451_00478
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Oxalobacter;
|
| CAZyme ID |
MGYG000003451_00478
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 583 |
|
65485.44 |
9.3793 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003451 |
1832891 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 95;
End: 1846
Strand: -
|
No EC number prediction in MGYG000003451_00478.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
24 |
511 |
1.5e-27 |
0.8407407407407408 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
4.02e-16 |
40 |
494 |
23 |
444 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| UAC69887.1
|
1.02e-21 |
37 |
579 |
72 |
621 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999881
|
0.000144
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
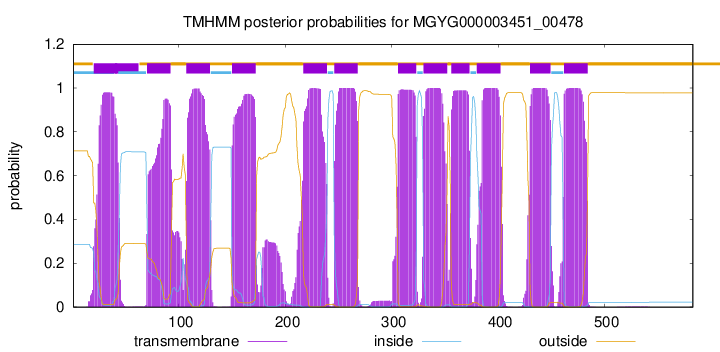
| start |
end |
| 20 |
42 |
| 70 |
92 |
| 107 |
129 |
| 150 |
172 |
| 217 |
239 |
| 246 |
268 |
| 306 |
323 |
| 330 |
352 |
| 356 |
373 |
| 380 |
402 |
| 430 |
449 |
| 462 |
484 |


