You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003479_00133
You are here: Home > Sequence: MGYG000003479_00133
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900548275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900548275 | |||||||||||
| CAZyme ID | MGYG000003479_00133 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3850; End: 5160 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 31 | 205 | 6.4e-20 | 0.39849624060150374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 4.37e-33 | 32 | 399 | 3 | 337 | Predicted dehydrogenase [General function prediction only]. |
| TIGR04380 | myo_inos_iolG | 1.13e-11 | 32 | 401 | 1 | 330 | inositol 2-dehydrogenase. All members of the seed alignment for this model are known or predicted inositol 2-dehydrogenase sequences co-clustered with other enzymes for catabolism of myo-inositol or closely related compounds. Inositol 2-dehydrogenase catalyzes the first step in inositol catabolism. Members of this family may vary somewhat in their ranges of acceptable substrates and some may act on analogs to myo-inositol rather than myo-inositol per se. [Energy metabolism, Sugars] |
| pfam01408 | GFO_IDH_MocA | 2.32e-10 | 33 | 172 | 1 | 116 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 5.73e-06 | 30 | 199 | 2 | 145 | putative oxidoreductase; Provisional |
| PRK10206 | PRK10206 | 1.65e-05 | 106 | 211 | 55 | 155 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQT66975.1 | 6.82e-94 | 2 | 436 | 12 | 425 |
| AOS46431.1 | 7.45e-92 | 2 | 436 | 10 | 432 |
| AWI08226.1 | 7.85e-91 | 1 | 436 | 4 | 430 |
| AGA27011.1 | 4.27e-90 | 3 | 436 | 9 | 429 |
| QEH36251.1 | 7.34e-90 | 2 | 436 | 8 | 424 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4N54_A | 6.96e-09 | 29 | 260 | 11 | 205 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 3CEA_A | 6.60e-07 | 33 | 302 | 9 | 236 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 3F4L_A | 4.83e-06 | 106 | 211 | 56 | 156 | Crystalstructure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_B Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_C Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_D Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_E Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_F Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01S58 | 6.81e-09 | 2 | 228 | 5 | 233 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| A0KYQ9 | 1.66e-08 | 2 | 200 | 5 | 206 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_2701 PE=3 SV=1 |
| Q0HH61 | 1.66e-08 | 2 | 200 | 5 | 206 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_2535 PE=3 SV=1 |
| Q0HTG8 | 1.66e-08 | 2 | 200 | 5 | 206 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-7) OX=60481 GN=Shewmr7_2602 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
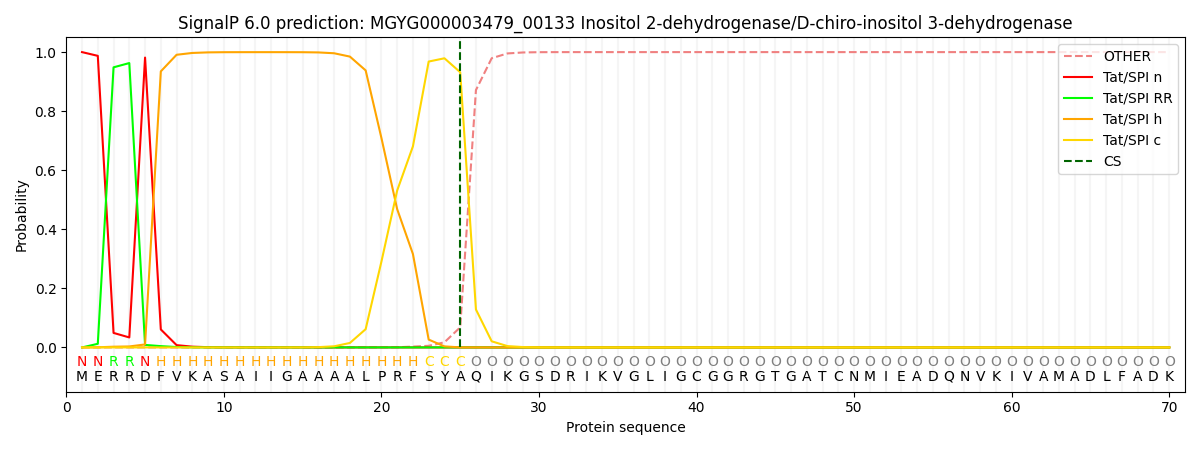
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999978 | 0.000000 | 0.000000 |
