You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003479_00324
You are here: Home > Sequence: MGYG000003479_00324
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900548275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900548275 | |||||||||||
| CAZyme ID | MGYG000003479_00324 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8414; End: 9985 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 84 | 473 | 1.2e-43 | 0.9603960396039604 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 7.46e-32 | 169 | 473 | 22 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 2.20e-24 | 105 | 473 | 24 | 308 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.55e-19 | 160 | 478 | 79 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA28189.1 | 1.25e-186 | 31 | 523 | 22 | 504 |
| QQZ02681.1 | 2.04e-178 | 30 | 516 | 18 | 488 |
| AHF92621.1 | 3.44e-174 | 35 | 522 | 16 | 490 |
| AVM47074.1 | 4.85e-173 | 22 | 523 | 3 | 496 |
| AWI10666.1 | 1.17e-156 | 37 | 523 | 1 | 480 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LPS_A | 6.90e-14 | 157 | 363 | 61 | 228 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2UWF_A | 7.18e-14 | 157 | 363 | 60 | 227 | ChainA, ALKALINE ACTIVE ENDOXYLANASE [Halalkalibacterium halodurans] |
| 3W27_A | 8.15e-14 | 156 | 476 | 59 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W28_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W29_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 7CPK_A | 1.77e-13 | 157 | 363 | 62 | 229 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 7CPL_A | 2.36e-13 | 157 | 363 | 62 | 229 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07528 | 1.21e-12 | 157 | 363 | 106 | 273 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| A1CHQ0 | 7.28e-11 | 166 | 473 | 91 | 313 | Probable endo-1,4-beta-xylanase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=xlnC PE=2 SV=1 |
| P07986 | 8.95e-11 | 245 | 475 | 127 | 351 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| C5J411 | 6.02e-10 | 254 | 473 | 130 | 323 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
| P23557 | 5.51e-09 | 168 | 347 | 30 | 164 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
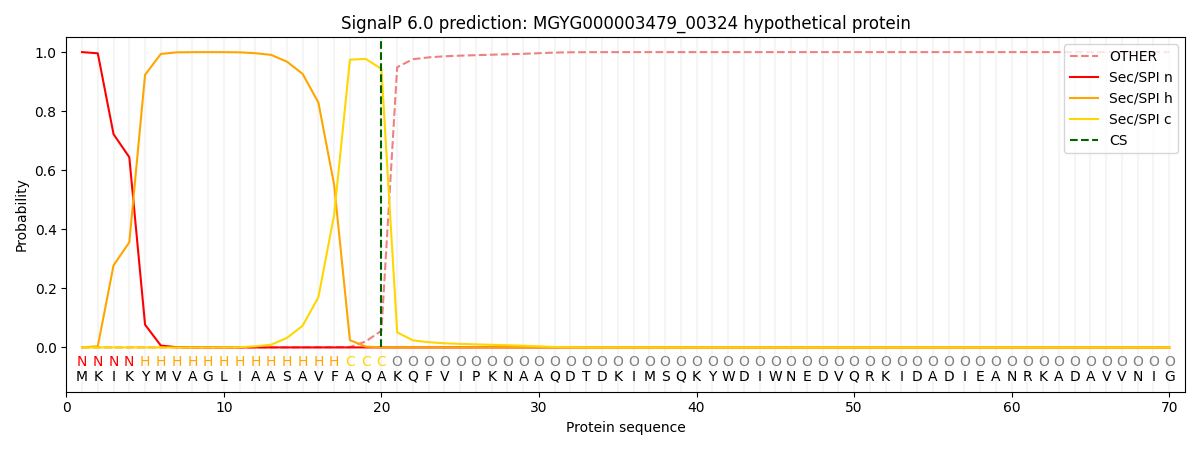
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000487 | 0.998782 | 0.000214 | 0.000178 | 0.000168 | 0.000152 |
