You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003493_01184
You are here: Home > Sequence: MGYG000003493_01184
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002299275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002299275 | |||||||||||
| CAZyme ID | MGYG000003493_01184 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18490; End: 21492 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 196 | 986 | 3e-261 | 0.9856527977044476 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 0.0 | 341 | 677 | 1 | 334 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| TIGR02985 | Sig70_bacteroi1 | 1.38e-26 | 9 | 161 | 9 | 161 | RNA polymerase sigma-70 factor, Bacteroides expansion family 1. This group of sigma factors are members of the sigma-70 family (TIGR02937) and are found primarily in the genus Bacteroides. This family appears to have resulted from a lineage-specific expansion as B. thetaiotaomicron VPI-5482, Bacteroides forsythus ATCC 43037, Bacteroides fragilis YCH46 and Bacteroides fragilis NCTC 9343 contain 25, 12, 24 and 23 members, respectively. There are currentlyonly two known members of this family outside of the Bacteroides, in Rhodopseudomonas and Bradyrhizobium. |
| pfam17829 | GH115_C | 4.74e-25 | 835 | 1000 | 6 | 168 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
| TIGR02937 | sigma70-ECF | 1.17e-18 | 5 | 161 | 1 | 158 | RNA polymerase sigma factor, sigma-70 family. This model encompasses all varieties of the sigma-70 type sigma factors including the ECF subfamily. A number of sigma factors have names with a different number than 70 (i.e. sigma-38), but in fact, all except for the Sigma-54 family (TIGR02395) are included within this family. Several Pfam models hit segments of these sequences including Sigma-70 region 2 (pfam04542) and Sigma-70, region 4 (pfam04545), but not always above their respective trusted cutoffs. |
| COG1595 | RpoE | 2.97e-17 | 1 | 162 | 11 | 176 | DNA-directed RNA polymerase specialized sigma subunit, sigma24 family [Transcription]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCG34758.1 | 0.0 | 193 | 1000 | 3 | 795 |
| QNT66545.1 | 0.0 | 174 | 997 | 19 | 849 |
| BCS85271.1 | 0.0 | 166 | 994 | 8 | 819 |
| ADE81569.1 | 0.0 | 166 | 996 | 11 | 816 |
| QVJ82146.1 | 0.0 | 166 | 996 | 10 | 815 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C90_A | 0.0 | 169 | 995 | 20 | 841 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 7PUG_A | 0.0 | 200 | 995 | 27 | 826 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 7PXQ_A | 0.0 | 200 | 995 | 26 | 825 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 4ZMH_A | 1.25e-170 | 187 | 828 | 2 | 643 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
| 6NPS_A | 8.73e-105 | 210 | 808 | 31 | 647 | Crystalstructure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
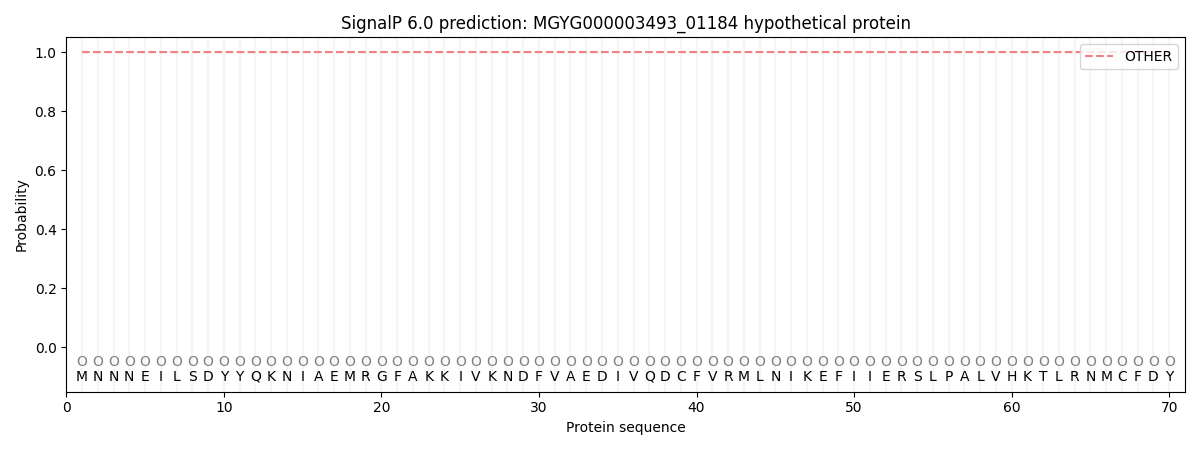
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000047 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
