You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003514_00278
You are here: Home > Sequence: MGYG000003514_00278
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fibrobacter_A intestinalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fibrobacterota; Fibrobacteria; Fibrobacterales; Fibrobacteraceae; Fibrobacter_A; Fibrobacter_A intestinalis | |||||||||||
| CAZyme ID | MGYG000003514_00278 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 77680; End: 79881 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 51 | 297 | 1.8e-79 | 0.9957805907172996 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 7.73e-48 | 52 | 300 | 4 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.13e-05 | 84 | 308 | 83 | 367 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| TIGR04183 | Por_Secre_tail | 8.09e-04 | 669 | 728 | 11 | 67 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL26286.1 | 1.47e-227 | 5 | 730 | 4 | 741 |
| ACX74827.1 | 1.47e-227 | 5 | 730 | 4 | 741 |
| QPB75689.1 | 5.53e-180 | 4 | 731 | 5 | 719 |
| QPB75699.1 | 1.41e-172 | 6 | 730 | 3 | 733 |
| AGS52465.1 | 5.91e-64 | 30 | 345 | 24 | 355 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EGZ_A | 2.09e-59 | 40 | 324 | 5 | 274 | ChainA, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_B Chain B, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_C Chain C, ENDOGLUCANASE Z [Dickeya chrysanthemi] |
| 1TVN_A | 2.07e-54 | 40 | 346 | 5 | 289 | Cellulasecel5G from Pseudoalteromonas haloplanktis, A family GH 5-2 enzyme [Pseudoalteromonas haloplanktis],1TVN_B Cellulase cel5G from Pseudoalteromonas haloplanktis, A family GH 5-2 enzyme [Pseudoalteromonas haloplanktis],1TVP_A Endoglucanase cel5G from Pseudoalteromonas haloplanktis in complex with cellobiose [Pseudoalteromonas haloplanktis],1TVP_B Endoglucanase cel5G from Pseudoalteromonas haloplanktis in complex with cellobiose [Pseudoalteromonas haloplanktis] |
| 5IHS_A | 6.40e-53 | 34 | 370 | 29 | 346 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 6GJF_A | 2.06e-43 | 33 | 347 | 6 | 302 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 1A3H_A | 6.89e-40 | 34 | 347 | 3 | 299 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07103 | 4.44e-57 | 40 | 324 | 48 | 317 | Endoglucanase Z OS=Dickeya dadantii (strain 3937) OX=198628 GN=celZ PE=1 SV=2 |
| O85465 | 3.58e-38 | 34 | 347 | 32 | 328 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P07983 | 3.68e-37 | 33 | 346 | 35 | 330 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P15704 | 1.87e-36 | 38 | 394 | 45 | 372 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 7.56e-36 | 33 | 342 | 35 | 326 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
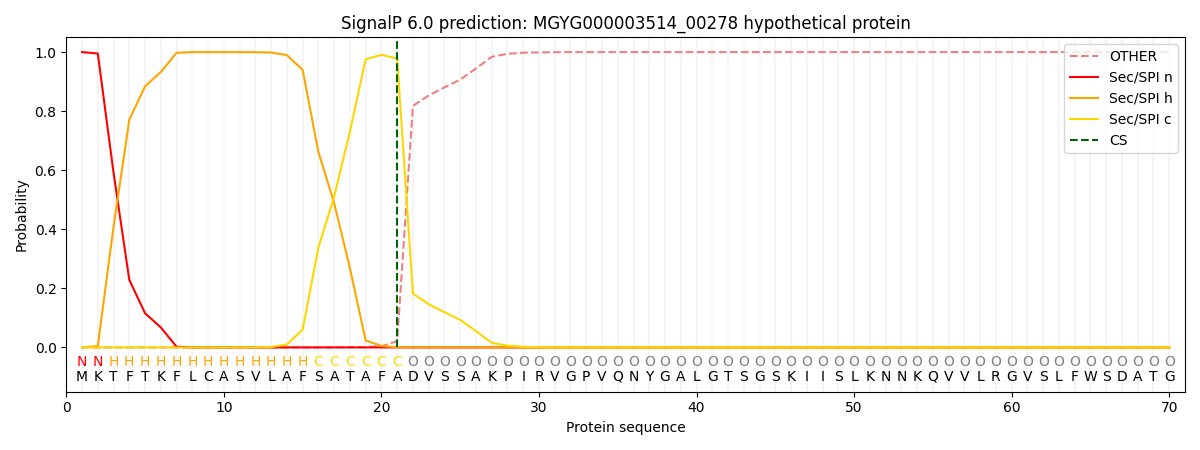
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999044 | 0.000181 | 0.000177 | 0.000169 | 0.000150 |
