You are browsing environment: HUMAN GUT
MGYG000003514_01196
Basic Information
help
Species
Fibrobacter_A intestinalis
Lineage
Bacteria; Fibrobacterota; Fibrobacteria; Fibrobacterales; Fibrobacteraceae; Fibrobacter_A; Fibrobacter_A intestinalis
CAZyme ID
MGYG000003514_01196
CAZy Family
CBM51
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
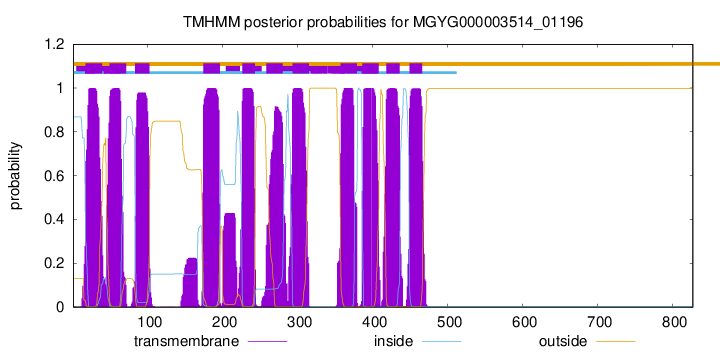
CGC
Molecular Weight
Isoelectric Point
827
93403.8
7.2239
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003514
2933446
MAG
Fiji
Oceania
Gene Location
Start: 5974;
End: 8457
Strand: -
No EC number prediction in MGYG000003514_01196.
Family
Start
End
Evalue
family coverage
CBM51
693
823
2e-35
0.9850746268656716
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam08305
NPCBM
1.04e-38
694
824
5
136
NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.)
more
smart00776
NPCBM
1.42e-21
694
824
7
145
This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins.
more
COG1807
ArnT
4.41e-06
162
463
76
368
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
B2UNU8
1.85e-11
704
826
81
209
Alpha-1,3-galactosidase B OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaB PE=3 SV=1
more
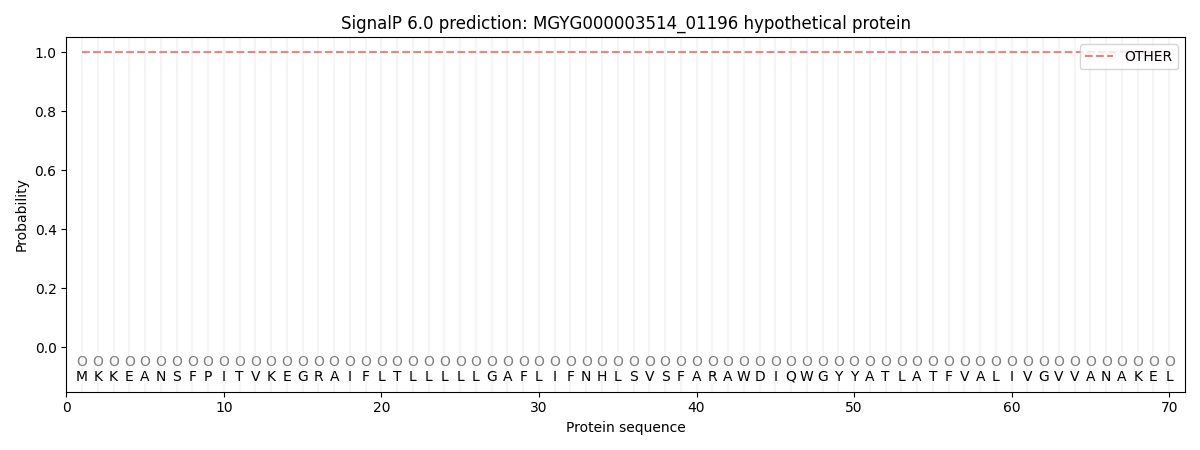
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000049
0.000001
0.000000
0.000000
0.000000
0.000000
start
end
17
39
49
71
83
102
174
196
226
243
258
280
293
315
357
379
386
408
418
436
449
466