You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003546_00972
You are here: Home > Sequence: MGYG000003546_00972
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1181 sp900769555 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Paludibacteraceae; UBA1181; UBA1181 sp900769555 | |||||||||||
| CAZyme ID | MGYG000003546_00972 | |||||||||||
| CAZy Family | GH44 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49857; End: 51752 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH44 | 25 | 543 | 1.2e-105 | 0.9941634241245136 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12891 | Glyco_hydro_44 | 3.78e-57 | 94 | 321 | 2 | 232 | Glycoside hydrolase family 44. This is a family of bacterial glycoside hydrolases formerly known as cellulase family J, and now known as Cel44A. It is one of the major enzymatic components of the cellulosome of Clostridium thermocellum strain F1 and of many other Firmicutes. |
| TIGR04183 | Por_Secre_tail | 1.06e-05 | 563 | 630 | 5 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| smart00813 | Alpha-L-AF_C | 6.20e-05 | 421 | 536 | 57 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam18962 | Por_Secre_tail | 3.82e-04 | 561 | 625 | 1 | 67 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBE16910.1 | 1.28e-185 | 38 | 630 | 36 | 636 |
| QRR00262.1 | 1.02e-184 | 28 | 626 | 20 | 617 |
| AEI47830.1 | 2.10e-183 | 21 | 626 | 11 | 619 |
| ACT92814.1 | 2.34e-181 | 28 | 618 | 27 | 618 |
| ADB40299.1 | 1.49e-176 | 25 | 631 | 18 | 620 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3FW6_A | 1.80e-24 | 27 | 487 | 19 | 487 | Crystalstructure of CelM2, a bifunctional glucanase-xylanase protein from a metagenome library [uncultured bacterium] |
| 3II1_A | 1.81e-24 | 27 | 487 | 19 | 487 | Structuralcharacterization of difunctional glucanase-xylanse CelM2 [uncultured bacterium] |
| 2YKK_A | 7.04e-14 | 25 | 485 | 7 | 467 | Structureof a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa],3ZQ9_A Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa] |
| 2YIH_A | 1.16e-12 | 25 | 485 | 7 | 467 | Structureof a Paenibacillus polymyxa Xyloglucanase from GH family 44 with Xyloglucan [Paenibacillus polymyxa],2YJQ_A Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa],2YJQ_B Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa] |
| 2E0P_A | 1.88e-11 | 19 | 322 | 1 | 287 | ChainA, Endoglucanase [Acetivibrio thermocellus],2E4T_A Chain A, Endoglucanase [Acetivibrio thermocellus],2EO7_A Chain A, Endoglucanase [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22533 | 2.86e-15 | 26 | 486 | 786 | 1255 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| P29719 | 1.89e-11 | 1 | 322 | 1 | 314 | Endoglucanase A OS=Paenibacillus lautus OX=1401 GN=celA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
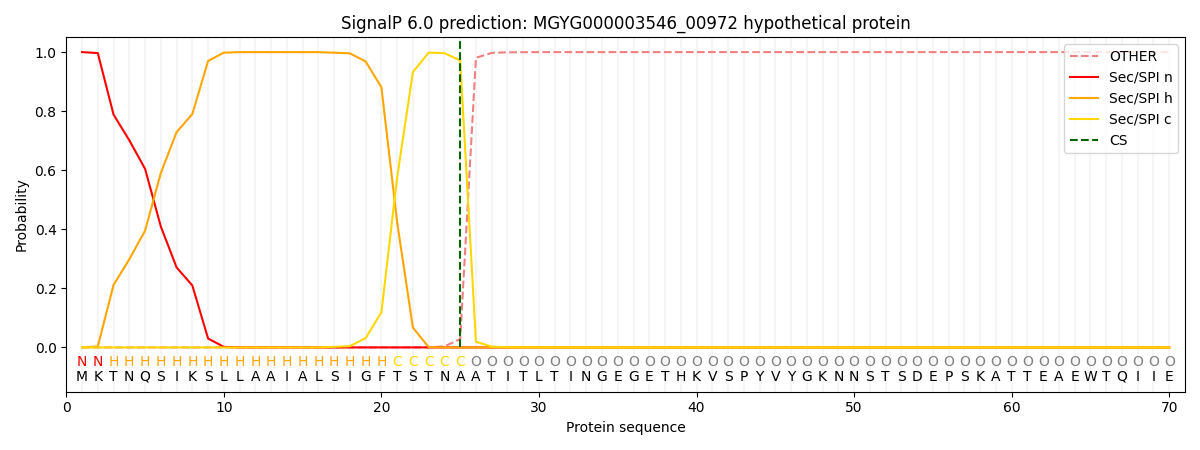
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998894 | 0.000190 | 0.000189 | 0.000179 | 0.000161 |
