You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003607_01525
You are here: Home > Sequence: MGYG000003607_01525
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM04593 sp900770655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; HGM04593; HGM04593 sp900770655 | |||||||||||
| CAZyme ID | MGYG000003607_01525 | |||||||||||
| CAZy Family | GT30 | |||||||||||
| CAZyme Description | 3-deoxy-D-manno-octulosonic acid transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3521; End: 4744 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT30 | 39 | 205 | 1.9e-44 | 0.9717514124293786 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1519 | KdtA | 3.07e-62 | 2 | 405 | 6 | 415 | 3-deoxy-D-manno-octulosonic-acid transferase [Cell wall/membrane/envelope biogenesis]. |
| PRK05749 | PRK05749 | 2.91e-53 | 6 | 405 | 13 | 417 | 3-deoxy-D-manno-octulosonic-acid transferase; Reviewed |
| pfam04413 | Glycos_transf_N | 2.40e-50 | 41 | 205 | 8 | 176 | 3-Deoxy-D-manno-octulosonic-acid transferase (kdotransferase). Members of this family transfer activated sugars to a variety of substrates, including glycogen, fructose-6-phosphate and lipopolysaccharides. Members of the family transfer UDP, ADP, GDP or CMP linked sugars. The Glycos_transf_N region is flanked at the N-terminus by a signal peptide and at the C-terminus by Glycos_transf_1 (pfam00534). The eukaryotic glycogen synthases may be distant members of this bacterial family. |
| cd03820 | GT4_AmsD-like | 0.004 | 109 | 387 | 77 | 333 | amylovoran biosynthesis glycosyltransferase AmsD and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. AmSD in Erwinia amylovora has been shown to be involved in the biosynthesis of amylovoran, the acidic exopolysaccharide acting as a virulence factor. This enzyme may be responsible for the formation of galactose alpha-1,6 linkages in amylovoran. |
| COG0707 | MurG | 0.006 | 135 | 405 | 107 | 351 | UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABR39163.1 | 8.23e-169 | 1 | 407 | 2 | 406 |
| QQY39259.1 | 8.23e-169 | 1 | 407 | 2 | 406 |
| ALK84072.1 | 1.66e-168 | 1 | 407 | 2 | 406 |
| QQY42562.1 | 1.66e-168 | 1 | 407 | 2 | 406 |
| QJR75178.1 | 1.66e-168 | 1 | 407 | 2 | 406 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XCI_A | 5.81e-29 | 20 | 372 | 15 | 342 | Membrane-embeddedmonofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCU_A Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O66663 | 1.55e-27 | 40 | 372 | 13 | 321 | 3-deoxy-D-manno-octulosonic acid transferase OS=Aquifex aeolicus (strain VF5) OX=224324 GN=kdtA PE=1 SV=1 |
| Q9PKI5 | 1.62e-20 | 41 | 396 | 49 | 411 | 3-deoxy-D-manno-octulosonic acid transferase OS=Chlamydia muridarum (strain MoPn / Nigg) OX=243161 GN=waaA PE=3 SV=1 |
| P0CE14 | 2.98e-20 | 42 | 360 | 51 | 378 | 3-deoxy-D-manno-octulosonic acid transferase OS=Chlamydia trachomatis (strain D/UW-3/Cx) OX=272561 GN=waaA PE=3 SV=1 |
| P0AC76 | 3.86e-20 | 42 | 405 | 46 | 416 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=waaA PE=3 SV=1 |
| P0AC77 | 3.86e-20 | 42 | 405 | 46 | 416 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O157:H7 OX=83334 GN=waaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
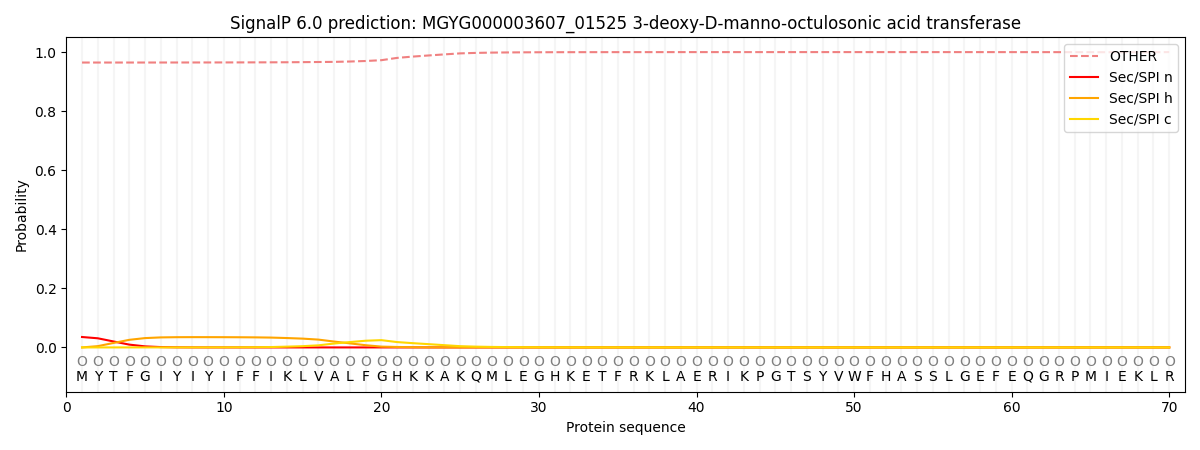
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.967076 | 0.032548 | 0.000171 | 0.000068 | 0.000043 | 0.000110 |
