You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003664_02320
You are here: Home > Sequence: MGYG000003664_02320
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
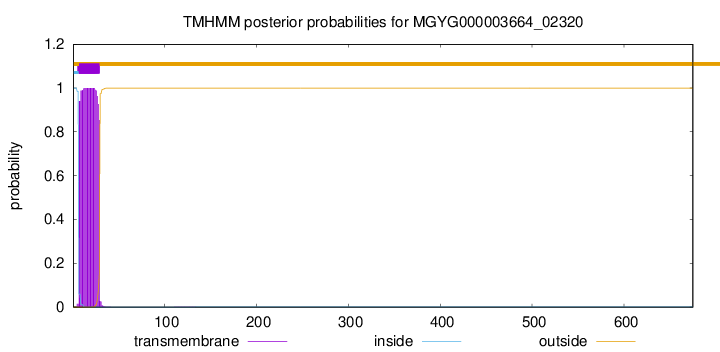
TMHMM annotations
Basic Information help
| Species | CAG-170 sp900751035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-170; CAG-170 sp900751035 | |||||||||||
| CAZyme ID | MGYG000003664_02320 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7277; End: 9304 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 46 | 370 | 8e-74 | 0.9837133550488599 |
| CBM42 | 541 | 671 | 1.6e-37 | 0.9779411764705882 |
| CBM13 | 398 | 540 | 1.2e-23 | 0.7180851063829787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05270 | AbfB | 2.92e-38 | 541 | 671 | 2 | 136 | Alpha-L-arabinofuranosidase B (ABFB) domain. This family consists of several fungal alpha-L-arabinofuranosidase B proteins. L-Arabinose is a constituent of plant-cell-wall poly-saccharides. It is found in a polymeric form in L-arabinan, in which the backbone is formed by 1,5-a- linked l-arabinose residues that can be branched via 1,2-a- and 1,3-a-linked l-arabinofuranose side chains. AbfB hydrolyzes 1,5-a, 1,3-a and 1,2-a linkages in both oligosaccharides and polysaccharides, which contain terminal non-reducing l-arabinofuranoses in side chains. |
| pfam14200 | RicinB_lectin_2 | 2.64e-21 | 435 | 520 | 3 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.19e-14 | 480 | 536 | 1 | 57 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.11e-13 | 395 | 474 | 10 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 2.04e-12 | 400 | 531 | 3 | 126 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD84758.1 | 2.91e-139 | 1 | 670 | 1 | 664 |
| QOT10786.1 | 8.17e-139 | 24 | 670 | 24 | 664 |
| CQR58317.1 | 2.79e-138 | 6 | 670 | 3 | 659 |
| QTH40928.1 | 3.85e-136 | 1 | 670 | 1 | 663 |
| SDS04433.1 | 3.81e-133 | 20 | 670 | 20 | 664 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3A71_A | 1.48e-29 | 44 | 385 | 22 | 348 | Highresolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
| 2W5N_A | 2.27e-24 | 39 | 343 | 22 | 318 | ChainA, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
| 2YDT_A | 2.27e-24 | 39 | 343 | 22 | 318 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
| 2YDP_A | 1.36e-23 | 39 | 343 | 22 | 318 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
| 2W5O_A | 2.46e-23 | 39 | 343 | 22 | 318 | ChainA, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8NK89 | 1.16e-07 | 558 | 631 | 372 | 446 | Alpha-L-arabinofuranosidase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfB PE=1 SV=1 |
| Q9C4B1 | 1.16e-07 | 558 | 631 | 372 | 446 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus awamori OX=105351 GN=abfB PE=1 SV=1 |
| A2R511 | 4.66e-07 | 558 | 631 | 372 | 446 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=abfB PE=3 SV=1 |
| P42255 | 4.66e-07 | 558 | 631 | 372 | 446 | Alpha-L-arabinofuranosidase B OS=Aspergillus niger OX=5061 GN=abfB PE=1 SV=1 |
| O74288 | 1.44e-06 | 541 | 633 | 360 | 461 | Alpha-L-arabinofuranosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=abfB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000215 | 0.999202 | 0.000160 | 0.000151 | 0.000142 | 0.000127 |