You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003669_01612
Basic Information
help
| Species |
UMGS1933 sp900772235
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; UMGS1933; UMGS1933 sp900772235
|
| CAZyme ID |
MGYG000003669_01612
|
| CAZy Family |
GT84 |
| CAZyme Description |
Cyclic beta-(1,2)-glucan synthase NdvB
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003669 |
1541608 |
MAG |
Peru |
South America |
|
| Gene Location |
Start: 13828;
End: 17184
Strand: -
|
MGYG000003669_01612 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P20471
|
8.29e-156 |
41 |
1107 |
54 |
1123 |
Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000034
|
0.000019
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
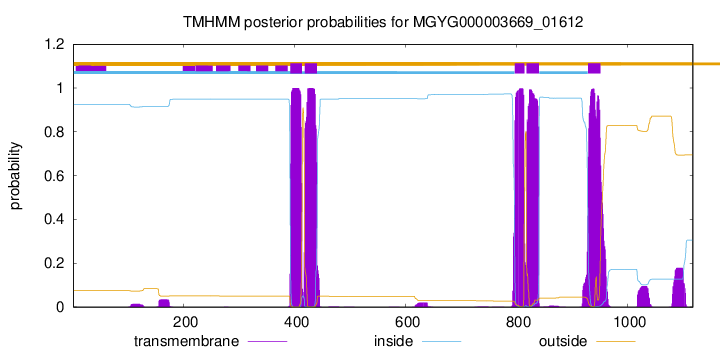
| start |
end |
| 392 |
413 |
| 418 |
440 |
| 797 |
814 |
| 818 |
840 |
| 929 |
951 |


