You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003687_05217
You are here: Home > Sequence: MGYG000003687_05217
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus polymyxa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus polymyxa | |||||||||||
| CAZyme ID | MGYG000003687_05217 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43619; End: 45100 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 61 | 293 | 1.7e-92 | 0.9915611814345991 |
| CBM59 | 348 | 492 | 2.7e-61 | 0.993103448275862 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.09e-58 | 77 | 299 | 19 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 7.13e-07 | 107 | 311 | 103 | 290 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUS26651.1 | 0.0 | 1 | 493 | 1 | 493 |
| QPK60378.1 | 0.0 | 1 | 493 | 1 | 493 |
| QPK55295.1 | 0.0 | 1 | 493 | 1 | 493 |
| QDA29283.1 | 0.0 | 1 | 493 | 1 | 493 |
| CCC85254.1 | 0.0 | 1 | 493 | 1 | 493 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EGZ_A | 2.90e-81 | 49 | 323 | 5 | 284 | ChainA, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_B Chain B, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_C Chain C, ENDOGLUCANASE Z [Dickeya chrysanthemi] |
| 1TVN_A | 1.67e-77 | 48 | 325 | 4 | 290 | Cellulasecel5G from Pseudoalteromonas haloplanktis, A family GH 5-2 enzyme [Pseudoalteromonas haloplanktis],1TVN_B Cellulase cel5G from Pseudoalteromonas haloplanktis, A family GH 5-2 enzyme [Pseudoalteromonas haloplanktis],1TVP_A Endoglucanase cel5G from Pseudoalteromonas haloplanktis in complex with cellobiose [Pseudoalteromonas haloplanktis],1TVP_B Endoglucanase cel5G from Pseudoalteromonas haloplanktis in complex with cellobiose [Pseudoalteromonas haloplanktis] |
| 6GJF_A | 5.30e-68 | 42 | 315 | 6 | 283 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 5IHS_A | 6.95e-68 | 43 | 332 | 29 | 304 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 4XZW_A | 1.50e-62 | 42 | 304 | 5 | 274 | Endo-glucanasechimera C10 [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07103 | 9.20e-79 | 49 | 323 | 48 | 327 | Endoglucanase Z OS=Dickeya dadantii (strain 3937) OX=198628 GN=celZ PE=1 SV=2 |
| P07983 | 2.21e-63 | 20 | 325 | 13 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P15704 | 4.95e-63 | 44 | 315 | 42 | 318 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 2.31e-62 | 20 | 315 | 13 | 312 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P23549 | 2.50e-60 | 20 | 315 | 13 | 312 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
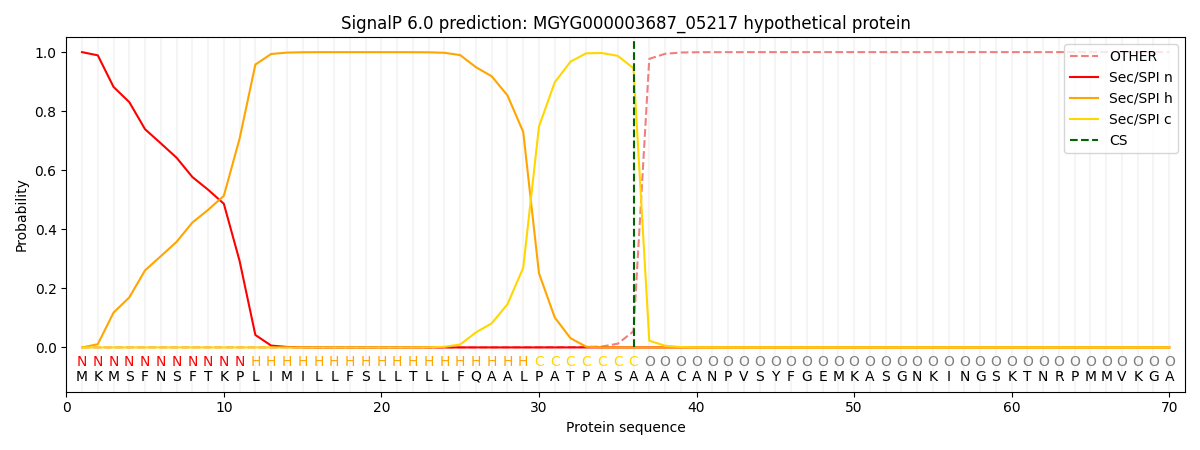
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000603 | 0.998504 | 0.000329 | 0.000192 | 0.000168 | 0.000165 |

