You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003689_01237
You are here: Home > Sequence: MGYG000003689_01237
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
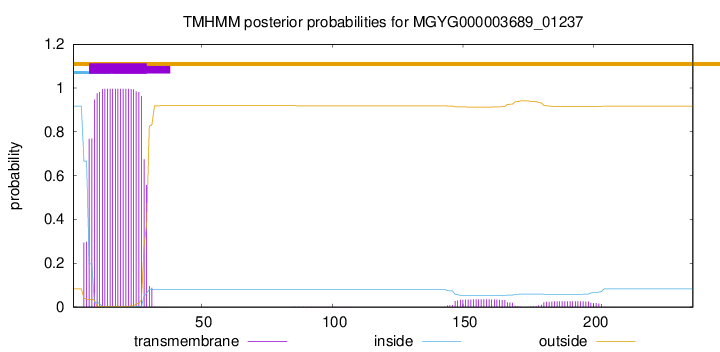
TMHMM annotations
Basic Information help
| Species | Faecalicoccus sp900546545 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Faecalicoccus; Faecalicoccus sp900546545 | |||||||||||
| CAZyme ID | MGYG000003689_01237 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4605; End: 5321 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 48 | 215 | 9.1e-64 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00912 | Transgly | 5.92e-84 | 52 | 215 | 14 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG0744 | MrcB | 3.69e-75 | 46 | 226 | 71 | 252 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 4.47e-61 | 1 | 227 | 1 | 243 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK00056 | mtgA | 4.69e-48 | 1 | 214 | 4 | 223 | monofunctional biosynthetic peptidoglycan transglycosylase; Provisional |
| TIGR02071 | PBP_1b | 1.18e-46 | 23 | 226 | 114 | 326 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUO38940.1 | 9.36e-81 | 2 | 225 | 4 | 228 |
| BAK43832.1 | 1.24e-74 | 2 | 229 | 20 | 244 |
| QJU22158.1 | 3.41e-74 | 27 | 238 | 29 | 234 |
| QRP41835.1 | 1.38e-73 | 27 | 238 | 29 | 234 |
| ASN93505.1 | 1.38e-73 | 27 | 238 | 29 | 234 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2OQO_A | 2.03e-38 | 53 | 224 | 22 | 193 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3NB6_A | 8.06e-38 | 53 | 224 | 22 | 193 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 5ZZK_A | 3.30e-35 | 42 | 234 | 48 | 246 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50] |
| 3VMQ_A | 4.99e-35 | 42 | 234 | 65 | 263 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMQ_B Chain B, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMR_A Chain A, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMS_A Chain A, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMS_B Chain B, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMT_A Chain A, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50],3VMT_B Chain B, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50] |
| 3HZS_A | 5.01e-35 | 42 | 233 | 12 | 209 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus MW2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38050 | 5.54e-44 | 54 | 225 | 70 | 241 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| O87579 | 1.90e-39 | 1 | 227 | 1 | 247 | Penicillin-binding protein 1A OS=Neisseria lactamica OX=486 GN=mrcA PE=3 SV=1 |
| P70997 | 2.45e-39 | 53 | 228 | 76 | 251 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| O86088 | 2.30e-38 | 48 | 227 | 65 | 247 | Penicillin-binding protein 1A OS=Neisseria cinerea OX=483 GN=mrcA PE=3 SV=1 |
| P0A0Z5 | 4.28e-38 | 48 | 227 | 65 | 247 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
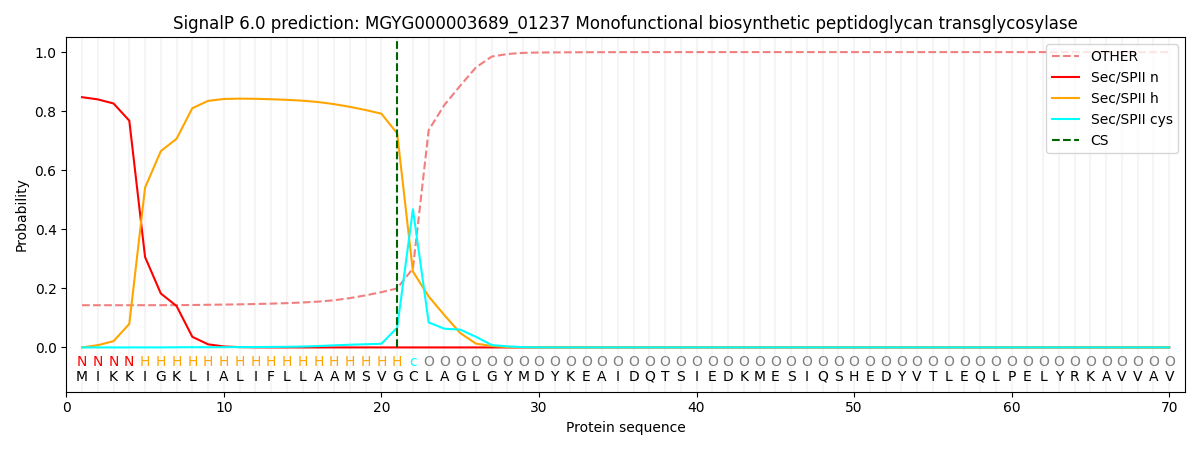
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.143270 | 0.009035 | 0.847527 | 0.000031 | 0.000031 | 0.000111 |