You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003693_00621
You are here: Home > Sequence: MGYG000003693_00621
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola plebeius_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola plebeius_A | |||||||||||
| CAZyme ID | MGYG000003693_00621 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14181; End: 16439 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 475 | 733 | 2.4e-40 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 5.29e-27 | 475 | 732 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 8.77e-27 | 475 | 734 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.98e-17 | 482 | 739 | 126 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 3.52e-11 | 53 | 135 | 3 | 85 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.29e-06 | 47 | 149 | 20 | 121 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRX63290.1 | 2.80e-288 | 19 | 738 | 22 | 731 |
| ANU56191.1 | 6.76e-276 | 1 | 736 | 1 | 743 |
| QQR18969.1 | 6.76e-276 | 1 | 736 | 1 | 743 |
| AXH21505.1 | 3.40e-260 | 11 | 739 | 12 | 746 |
| EDV05072.1 | 3.40e-260 | 11 | 739 | 12 | 746 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 1.92e-20 | 152 | 296 | 1 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 3.00e-18 | 162 | 293 | 5 | 141 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 4XUY_A | 8.51e-11 | 476 | 736 | 102 | 302 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 6LPS_A | 1.61e-08 | 461 | 651 | 100 | 261 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2F8Q_A | 3.80e-08 | 461 | 651 | 98 | 263 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O94163 | 2.31e-12 | 476 | 736 | 125 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| Q4JHP5 | 9.85e-12 | 476 | 736 | 126 | 326 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus OX=33178 GN=xlnC PE=2 SV=1 |
| Q0CBM8 | 1.32e-11 | 476 | 736 | 126 | 326 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnC PE=2 SV=2 |
| Q96VB6 | 7.35e-11 | 476 | 736 | 123 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| O59859 | 2.43e-10 | 476 | 736 | 127 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
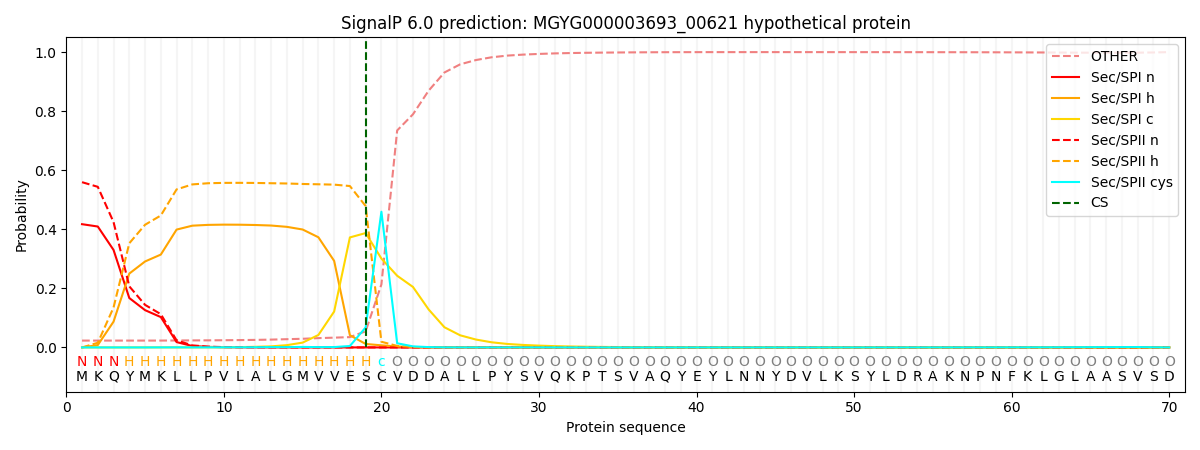
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.027247 | 0.407719 | 0.564345 | 0.000188 | 0.000214 | 0.000274 |
