You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003701_02405
You are here: Home > Sequence: MGYG000003701_02405
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp003480915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp003480915 | |||||||||||
| CAZyme ID | MGYG000003701_02405 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 291; End: 1661 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 63 | 227 | 1.5e-21 | 0.39598997493734334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 2.01e-38 | 60 | 418 | 1 | 342 | Predicted dehydrogenase [General function prediction only]. |
| TIGR04380 | myo_inos_iolG | 2.44e-19 | 62 | 290 | 1 | 198 | inositol 2-dehydrogenase. All members of the seed alignment for this model are known or predicted inositol 2-dehydrogenase sequences co-clustered with other enzymes for catabolism of myo-inositol or closely related compounds. Inositol 2-dehydrogenase catalyzes the first step in inositol catabolism. Members of this family may vary somewhat in their ranges of acceptable substrates and some may act on analogs to myo-inositol rather than myo-inositol per se. [Energy metabolism, Sugars] |
| pfam01408 | GFO_IDH_MocA | 1.73e-12 | 63 | 193 | 1 | 116 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK10206 | PRK10206 | 2.32e-10 | 131 | 337 | 59 | 242 | putative oxidoreductase; Provisional |
| PRK11579 | PRK11579 | 5.11e-09 | 136 | 267 | 64 | 186 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCM55249.1 | 2.85e-278 | 1 | 455 | 1 | 454 |
| QRX64436.1 | 3.06e-265 | 1 | 451 | 1 | 449 |
| SCM56422.1 | 8.11e-148 | 1 | 436 | 7 | 434 |
| BBD44694.1 | 4.91e-144 | 1 | 436 | 7 | 434 |
| SCD21857.1 | 5.42e-144 | 3 | 434 | 10 | 436 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CEA_A | 1.22e-13 | 61 | 348 | 7 | 284 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 4N54_A | 3.06e-13 | 61 | 350 | 13 | 280 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4MKX_A | 3.13e-13 | 61 | 350 | 16 | 283 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4HKT_A | 3.80e-10 | 63 | 344 | 4 | 255 | Crystalstructure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_B Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_C Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_D Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021] |
| 3Q2I_A | 7.61e-09 | 61 | 360 | 12 | 279 | Crystalstructure of the WlbA dehydrognase from Chromobactrium violaceum in complex with NADH and UDP-GlcNAcA at 1.50 A resolution [Chromobacterium violaceum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O05389 | 3.72e-12 | 61 | 382 | 3 | 298 | Uncharacterized oxidoreductase YrbE OS=Bacillus subtilis (strain 168) OX=224308 GN=yrbE PE=3 SV=2 |
| O68965 | 4.86e-10 | 63 | 344 | 3 | 254 | Inositol 2-dehydrogenase OS=Rhizobium meliloti (strain 1021) OX=266834 GN=idhA PE=1 SV=2 |
| A1S4U5 | 1.42e-09 | 21 | 237 | 17 | 221 | Glycosyl hydrolase family 109 protein OS=Shewanella amazonensis (strain ATCC BAA-1098 / SB2B) OX=326297 GN=Sama_1194 PE=3 SV=1 |
| P0C863 | 3.38e-09 | 63 | 171 | 54 | 159 | Glycosyl hydrolase family 109 protein 1 OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=BF0931 PE=3 SV=1 |
| Q5LGZ0 | 3.38e-09 | 63 | 171 | 54 | 159 | Glycosyl hydrolase family 109 protein 1 OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=BF0853 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
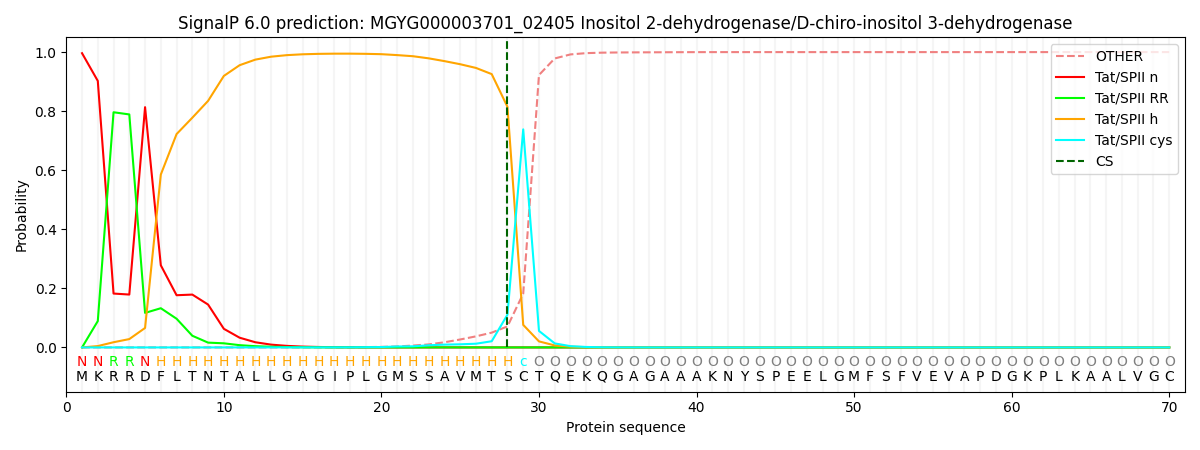
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000000 | 0.001786 | 0.001930 | 0.996274 | 0.000000 |
