You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003740_01181
You are here: Home > Sequence: MGYG000003740_01181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
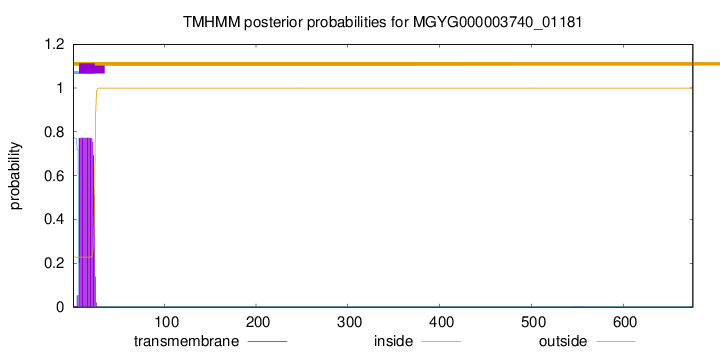
TMHMM annotations
Basic Information help
| Species | Ezakiella coagulans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Peptoniphilaceae; Ezakiella; Ezakiella coagulans | |||||||||||
| CAZyme ID | MGYG000003740_01181 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15834; End: 17864 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 530 | 668 | 1.2e-27 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 8.29e-38 | 521 | 675 | 43 | 191 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| NF038016 | sporang_Gsm | 7.94e-29 | 515 | 672 | 151 | 312 | sporangiospore maturation cell wall hydrolase GsmA. The peptidoglycan-hydrolyzing enzyme GsmA occurs in some sporangia-forming members of the Actinobacteria, such as Actinoplanes missouriensis, and is required for proper separation of spores. GsmA proteins have one or two SH3 domains N-terminal to the hydrolase domain. |
| pfam07833 | Cu_amine_oxidN1 | 3.12e-23 | 45 | 138 | 1 | 93 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| PRK05684 | flgJ | 7.28e-22 | 524 | 663 | 155 | 294 | flagellar assembly peptidoglycan hydrolase FlgJ. |
| PRK12713 | flgJ | 7.20e-21 | 524 | 663 | 184 | 323 | flagellar rod assembly protein/muramidase FlgJ; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY79269.1 | 5.84e-136 | 22 | 675 | 29 | 702 |
| QAT62865.1 | 7.96e-136 | 18 | 675 | 50 | 725 |
| VDN47006.1 | 7.48e-132 | 28 | 675 | 34 | 697 |
| AHM58209.1 | 1.87e-130 | 9 | 675 | 18 | 714 |
| QRN84622.1 | 2.39e-129 | 23 | 675 | 25 | 695 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VWO_A | 1.32e-14 | 524 | 670 | 4 | 150 | Crystalstructure of peptidoglycan hydrolase mutant from Sphingomonas sp. A1 [Sphingomonas sp. A1] |
| 2ZYC_A | 1.63e-14 | 524 | 670 | 5 | 151 | ChainA, Peptidoglycan hydrolase FlgJ [Sphingomonas sp. A1] |
| 3K3T_A | 1.03e-13 | 524 | 670 | 5 | 151 | E185Amutant of peptidoglycan hydrolase from Sphingomonas sp. A1 [Sphingomonas sp. A1] |
| 5DN5_A | 3.82e-13 | 524 | 663 | 6 | 145 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5DN4_A | 5.33e-13 | 524 | 663 | 6 | 145 | Structureof the glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9X9J3 | 2.63e-14 | 492 | 662 | 136 | 301 | Peptidoglycan hydrolase FlgJ OS=Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633) OX=223926 GN=flgJ PE=3 SV=1 |
| Q9KQ15 | 2.64e-14 | 521 | 670 | 161 | 313 | Peptidoglycan hydrolase FlgJ OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=flgJ PE=3 SV=2 |
| O32083 | 2.17e-12 | 524 | 675 | 51 | 201 | Exo-glucosaminidase LytG OS=Bacillus subtilis (strain 168) OX=224308 GN=lytG PE=1 SV=1 |
| P58231 | 2.31e-12 | 524 | 663 | 152 | 291 | Peptidoglycan hydrolase FlgJ OS=Escherichia coli O157:H7 OX=83334 GN=flgJ PE=3 SV=1 |
| P75942 | 2.31e-12 | 524 | 663 | 152 | 291 | Peptidoglycan hydrolase FlgJ OS=Escherichia coli (strain K12) OX=83333 GN=flgJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
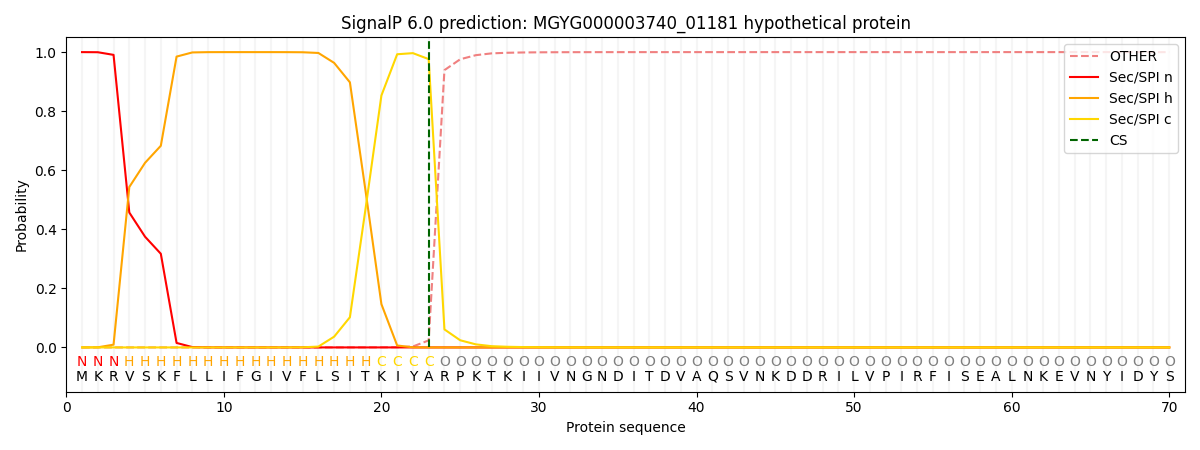
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000412 | 0.998823 | 0.000274 | 0.000158 | 0.000155 | 0.000145 |