You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003744_01431
You are here: Home > Sequence: MGYG000003744_01431
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
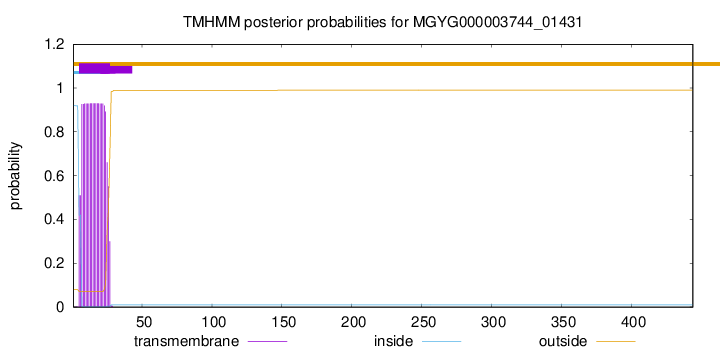
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; | |||||||||||
| CAZyme ID | MGYG000003744_01431 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1655; End: 2989 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 56 | 243 | 8.1e-20 | 0.45614035087719296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.29e-35 | 54 | 412 | 1 | 342 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 9.15e-11 | 57 | 179 | 1 | 112 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK10206 | PRK10206 | 8.00e-04 | 127 | 213 | 62 | 145 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23801.1 | 2.40e-241 | 1 | 444 | 7 | 453 |
| BCG53919.1 | 1.11e-227 | 1 | 444 | 7 | 459 |
| CDN31231.1 | 1.74e-227 | 1 | 444 | 5 | 454 |
| QUT52998.1 | 4.55e-222 | 1 | 441 | 7 | 454 |
| BBK93797.1 | 1.30e-221 | 1 | 441 | 7 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Z3B_B | 1.36e-08 | 102 | 301 | 34 | 221 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
| 6Z3B_A | 1.39e-08 | 102 | 301 | 36 | 223 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
| 6Z3C_AAA | 1.42e-08 | 102 | 301 | 42 | 229 | ChainAAA, Gfo/Idh/MocA family oxidoreductase [[Ruminococcus] gnavus],6Z3C_BBB Chain BBB, Gfo/Idh/MocA family oxidoreductase [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RK81 | 1.09e-10 | 1 | 217 | 7 | 214 | Glycosyl hydrolase family 109 protein OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO0529 PE=3 SV=1 |
| A8H2K3 | 5.32e-08 | 1 | 226 | 3 | 217 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
| A4Y8C8 | 7.10e-08 | 1 | 226 | 4 | 219 | Glycosyl hydrolase family 109 protein OS=Shewanella putrefaciens (strain CN-32 / ATCC BAA-453) OX=319224 GN=Sputcn32_2490 PE=3 SV=1 |
| A1RI61 | 1.25e-07 | 1 | 226 | 4 | 219 | Glycosyl hydrolase family 109 protein OS=Shewanella sp. (strain W3-18-1) OX=351745 GN=Sputw3181_1518 PE=3 SV=1 |
| Q8ECL7 | 2.91e-07 | 1 | 226 | 4 | 219 | Alpha-N-acetylgalactosaminidase OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=nagA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
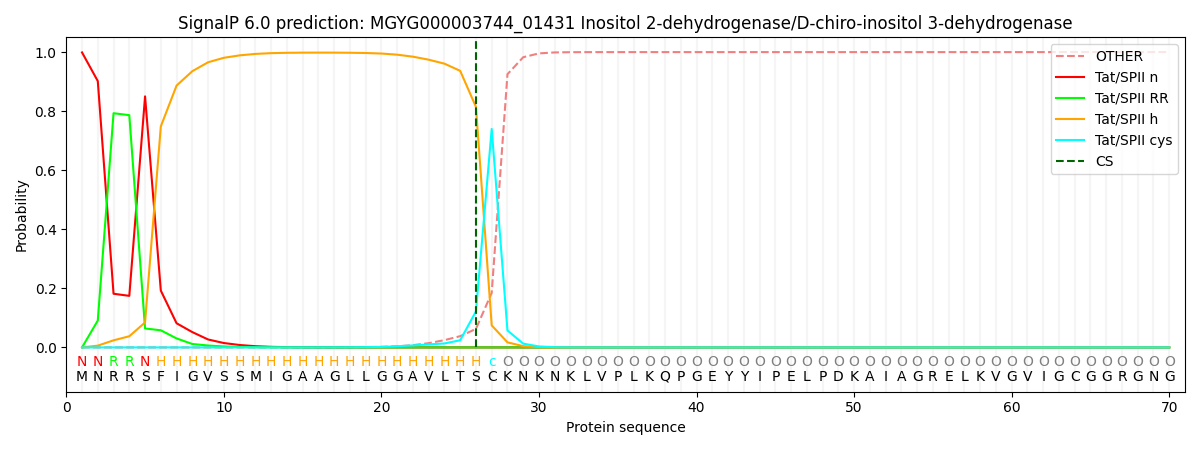
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000020 | 0.001123 | 0.998859 | 0.000000 |