You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003833_00768
You are here: Home > Sequence: MGYG000003833_00768
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; WRGP01; | |||||||||||
| CAZyme ID | MGYG000003833_00768 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4567; End: 6549 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 145 | 354 | 1.8e-57 | 0.9166666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 1.72e-83 | 15 | 658 | 5 | 648 | beta-glucosidase BglX. |
| COG1472 | BglX | 3.18e-58 | 85 | 496 | 1 | 377 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 2.80e-46 | 86 | 416 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 2.55e-43 | 454 | 658 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PLN03080 | PLN03080 | 8.53e-34 | 76 | 658 | 51 | 633 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE67433.1 | 0.0 | 1 | 659 | 1 | 659 |
| QTE67436.1 | 6.93e-119 | 7 | 656 | 4 | 650 |
| QSG05048.1 | 1.19e-105 | 67 | 660 | 10 | 629 |
| QSG12957.1 | 9.58e-104 | 67 | 660 | 10 | 629 |
| QIB74454.1 | 2.86e-100 | 80 | 658 | 21 | 624 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z9S_A | 1.21e-67 | 74 | 658 | 24 | 663 | Functionaland Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum],5Z9S_B Functional and Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum] |
| 5M6G_A | 5.01e-66 | 67 | 658 | 40 | 614 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 5XXL_A | 8.88e-66 | 72 | 658 | 8 | 635 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 5JP0_A | 3.83e-65 | 75 | 658 | 14 | 637 | Bacteroidesovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus],5JP0_B Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus] |
| 6R5I_A | 4.06e-65 | 142 | 660 | 62 | 618 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 1.56e-77 | 36 | 658 | 27 | 654 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q5BCC6 | 5.51e-74 | 63 | 660 | 34 | 617 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| Q2UFP8 | 2.31e-72 | 52 | 658 | 33 | 626 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 7.94e-72 | 66 | 658 | 41 | 622 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| A7LXU3 | 2.56e-64 | 75 | 658 | 36 | 659 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
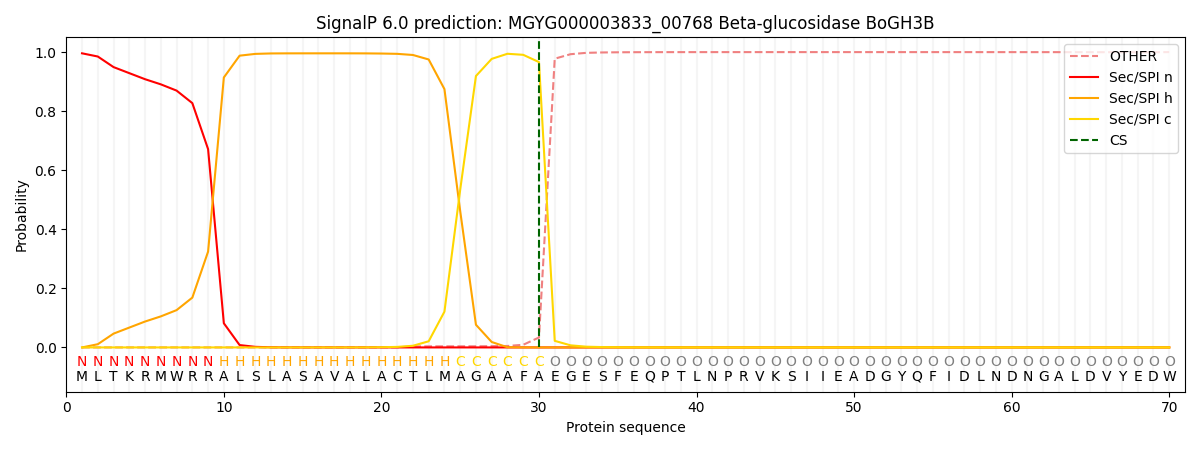
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000483 | 0.993453 | 0.005124 | 0.000491 | 0.000236 | 0.000184 |

