You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003893_00737
You are here: Home > Sequence: MGYG000003893_00737
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
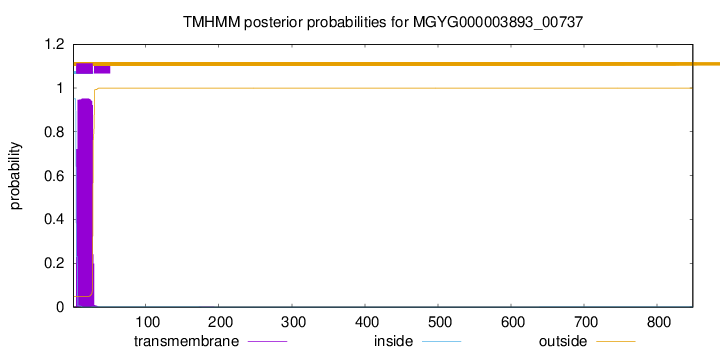
TMHMM annotations
Basic Information help
| Species | Prevotella multisaccharivorax | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella multisaccharivorax | |||||||||||
| CAZyme ID | MGYG000003893_00737 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Exo-beta-D-glucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27346; End: 29895 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 42 | 676 | 1.1e-58 | 0.6316489361702128 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18368 | Ig_GlcNase | 6.00e-30 | 735 | 841 | 1 | 104 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| COG3250 | LacZ | 8.66e-21 | 13 | 459 | 2 | 428 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.16e-07 | 88 | 459 | 77 | 444 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 9.13e-07 | 186 | 298 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK09525 | lacZ | 4.21e-04 | 171 | 323 | 208 | 354 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT89889.1 | 1.70e-293 | 23 | 848 | 29 | 871 |
| ALJ58991.1 | 3.91e-292 | 23 | 848 | 29 | 871 |
| QVJ81627.1 | 1.87e-270 | 30 | 836 | 35 | 817 |
| ADE83122.1 | 2.65e-270 | 30 | 836 | 35 | 817 |
| ATL49270.1 | 6.27e-266 | 36 | 849 | 63 | 871 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VZS_A | 6.84e-131 | 30 | 840 | 79 | 890 | ChitosanProduct complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
| 2VZO_A | 2.61e-130 | 30 | 840 | 79 | 890 | Crystalstructure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZT_A | 5.10e-130 | 30 | 840 | 79 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
| 2VZU_A | 1.45e-128 | 30 | 840 | 79 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
| 6BYE_A | 2.94e-23 | 77 | 685 | 71 | 726 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82NR8 | 3.15e-134 | 31 | 840 | 76 | 895 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
| Q56F26 | 3.75e-130 | 30 | 840 | 79 | 890 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
| D4AUH1 | 6.41e-130 | 14 | 842 | 51 | 878 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
| Q4R1C4 | 1.27e-112 | 45 | 840 | 72 | 872 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
| C0LRA7 | 2.80e-105 | 64 | 840 | 91 | 870 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
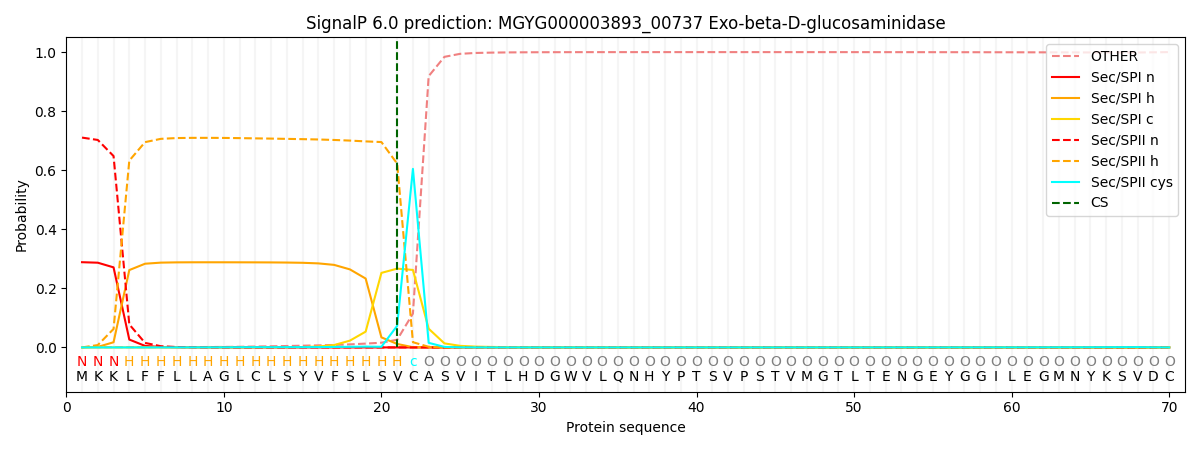
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002191 | 0.282160 | 0.715228 | 0.000148 | 0.000132 | 0.000151 |