You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003957_00793
You are here: Home > Sequence: MGYG000003957_00793
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp014287585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp014287585 | |||||||||||
| CAZyme ID | MGYG000003957_00793 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41261; End: 42562 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 80 | 383 | 6.9e-48 | 0.9504950495049505 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 8.48e-41 | 111 | 380 | 3 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 5.63e-37 | 90 | 383 | 23 | 308 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.71e-32 | 111 | 383 | 69 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEW21052.1 | 1.27e-181 | 25 | 432 | 25 | 434 |
| BAR51127.1 | 1.27e-181 | 25 | 432 | 25 | 434 |
| BAR48814.1 | 1.27e-181 | 25 | 432 | 25 | 434 |
| QGA25900.1 | 5.70e-181 | 19 | 433 | 23 | 440 |
| QQZ02681.1 | 8.49e-125 | 19 | 432 | 18 | 494 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 4.66e-22 | 46 | 432 | 52 | 411 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1XYZ_A | 4.87e-20 | 137 | 383 | 90 | 340 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 1VBR_A | 2.01e-18 | 111 | 390 | 53 | 323 | Crystalstructure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
| 3NIY_A | 3.16e-18 | 111 | 390 | 69 | 339 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
| 5AY7_A | 2.97e-17 | 120 | 383 | 61 | 343 | Apsychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5AY7_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5D4Y_A A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples],5D4Y_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 1.37e-18 | 137 | 383 | 580 | 830 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| A3DH97 | 3.06e-18 | 43 | 419 | 385 | 733 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| Q60041 | 4.50e-17 | 111 | 392 | 72 | 344 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| A0A1P8B8F8 | 4.28e-15 | 43 | 414 | 178 | 533 | Endo-1,4-beta-xylanase 5 OS=Arabidopsis thaliana OX=3702 GN=XYN5 PE=3 SV=1 |
| P23360 | 2.01e-14 | 111 | 383 | 77 | 323 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
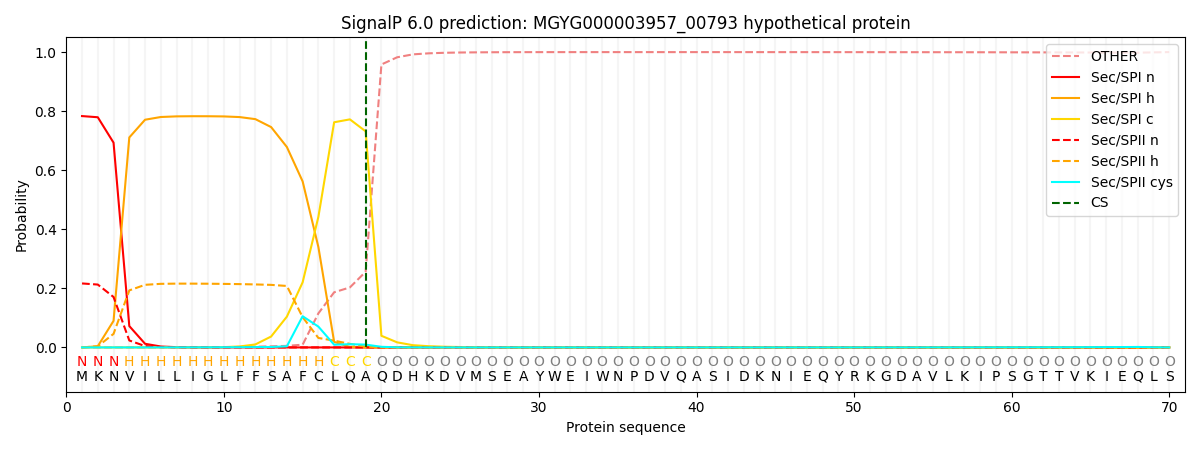
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000621 | 0.772581 | 0.225965 | 0.000280 | 0.000303 | 0.000255 |
